Key points
- The number of highly aggressive Ascochyta rabiei isolates on chickpea variety PBA Seamer (PBR) has risen from four per cent in 2016 to 48 per cent in 2020, dropping its industry classification from ‘resistant’ to ‘moderately susceptible’
- The trend appears to be occurring for other ‘resistant’ and ‘moderately resistant’ varieties and is being investigated by Griffith University researchers
- An online dashboard has been created to help inform disease management strategies and identify trends and patterns affecting the emergence of highly aggressive isolates
A new online dashboard is playing an important role in investigating Australian Ascochyta rabiei populations and their ability to adapt to disease management strategies.
Developed by the GRDC-supported chickpea Ascochyta blight research program team at Brisbane’s Griffith University, the dashboard aims to support informed disease management strategies and to identify trends and patterns affecting the emergence of highly aggressive isolates.
It is one part of the research program’s continued and deeper investigation into why highly aggressive Ascochyta rabiei isolates are increasing.
Speaking at the GRDC Updates, Griffith University’s Dr Ido Bar said that in the space of just four years, the number of highly aggressive A. rabiei isolates on the ‘resistant’ chickpea variety PBA Seamer (PBR) had risen from four per cent in 2016 to 48 per cent in 2020. It has forced the variety to be downgraded from ‘resistant’ to ‘moderately susceptible’.
Over the past eight years, researchers have established a comprehensive isolate database, and each year they determine isolates’ aggressiveness against chickpea genotypes.
Dashboard
The dashboard is the latest research tool developed to track this development. It provides a spatio-temporal overview of 1096 isolates that were collected between 2013 and 2021, along with specific details of their collection metadata.
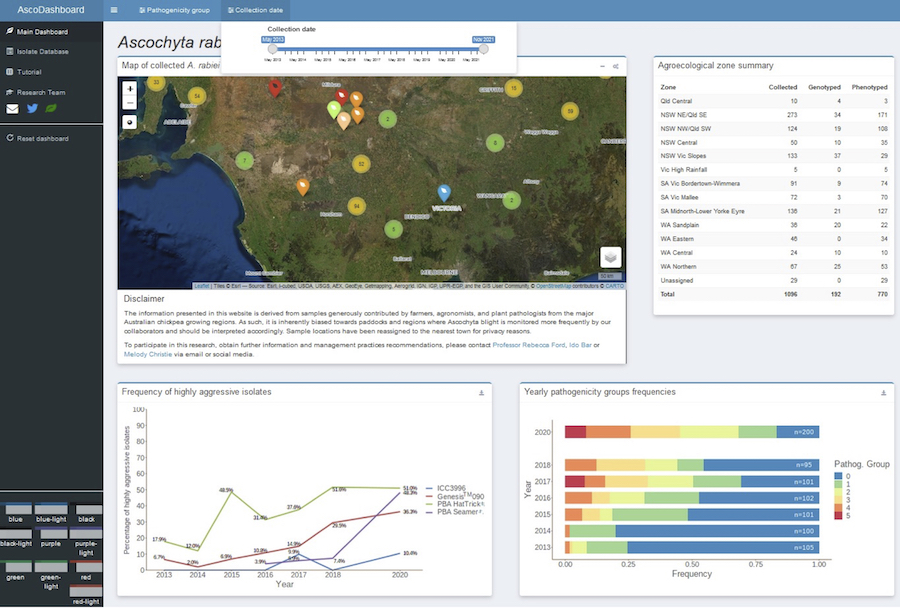
A screenshot of the Ascochyta Dashboard developed by the team at Griffith University. It shows the spatial distribution of Ascochyta rabiei isolates, summary tables, graphs and interface controls. See ASCO Dashboard.
It also details the phenotypic assessment and summary plots of 806 isolates that were phenotyped on a differential host set for their aggressiveness between 2013 and 2020. See See ASCO Dashboard.
The dashboard is available to growers, researchers and agronomists and can assist in disease management, such as cultivar selection and fungicide application, by providing information on potential hotspots around the country and cultivars that could induce the emergence of highly aggressive isolates.
Genetic understanding
Previous research has shown that A. rabiei in Australia appears to be clonal. This means offspring are identical to their parent (apart from point mutations), leading to a limited genetic diversity. This would usually suggest limited adaptation potential, but this is not the case.
Dr Bar said the population seems to contain sufficient existing diversity to undergo spontaneous mutation events to rapidly adapt to and overcome host resistance sources.
“This is evident by the detection of severe disease symptoms on the formerly released ‘resistant’ cultivars, such as Genesis™ 090, PBA Seamer (PBR) and ICC3996, the resistance source for many commercial varieties,” he said.
The proportion of isolates highly aggressive on PBA HatTrick (PBR) rose from 18 per cent in 2013 to almost 50 per cent in just one year. Similarly, a higher frequency of isolates able to cause severe disease on Genesis™ 090 was observed, up from 11 per cent in 2016 to 36 per cent in 2020.
Dr Bar said this was particularly concerning given that PBA HatTrick (PBR) and Genesis 090 are broadly sown cultivars in northern and southern chickpea regions, respectively.

Griffith University’s Dr Ido Bar recently spoke at GRDC Updates about research to understand Ascochyta rabiei’s increasing aggressiveness. Photo: Courtesy Ido Bar
To better understand this, between 2013 and 2018 the team collected 279 isolates from across the major growing regions for high-resolution genetic analysis.
This helped discover clusters of isolates with a similar genetic background that showed high proportions of highly aggressive isolates, which potentially pose serious risks to the industry, Dr Bar said.
Isolates in these clusters are seven to 13 times more likely to be highly aggressive than isolates with a different genetic background.
“We identified two likely drivers that contribute to the emergence of new high-risk isolates: either local micro-evolution of isolates that can lead to adaptation of highly aggressive isolates, or cross-region gene flow, likely driven by human activities,” he said.
With both mechanisms posing ongoing risks, rapidly identifying emerging isolates and their genetic make-up is crucial to be able to manage the risk and control the disease effectively in an event of an outbreak.
The Ascochyta team includes Griffith University’s Melody Christie, Professor Rebecca Ford, Dr Ido Bar and Dr Prabhakaran Sambasivam, as well as the University of Melbourne’s Dr Niloofar Vaghefi and Dr Adam Sparks from Western Australia’s Department of Primary Industries and Regional Development.
More information: Ido Bar, 0435 718 770, i.bar@griffith.edu.au
Read also: Plan of attack for aggressive ascochyta isolates.