Rhizobia as an inoculant on legume crops saves the industry an estimated $3.5 billion each year in applied nitrogen costs. Now, a new facility is taking the inoculant development pipeline to a whole new level
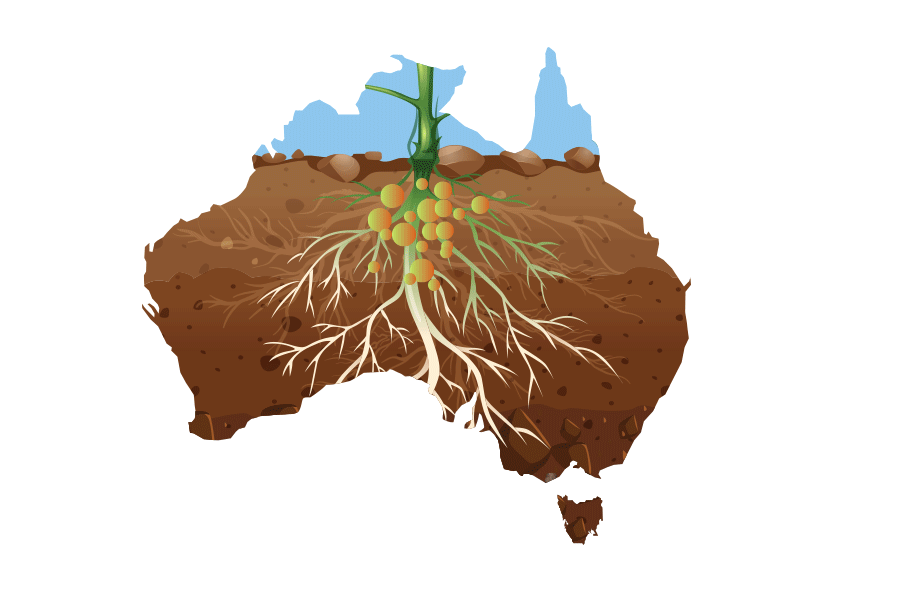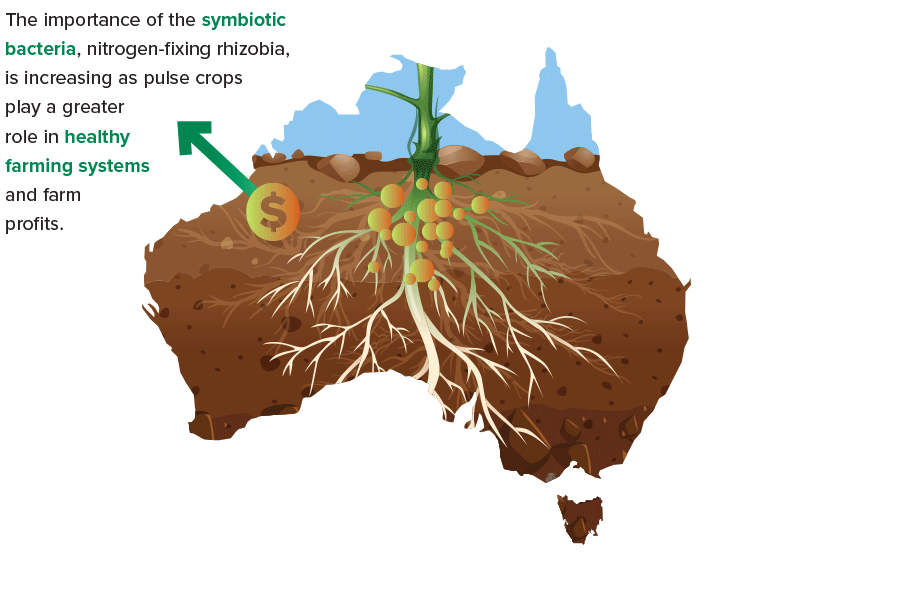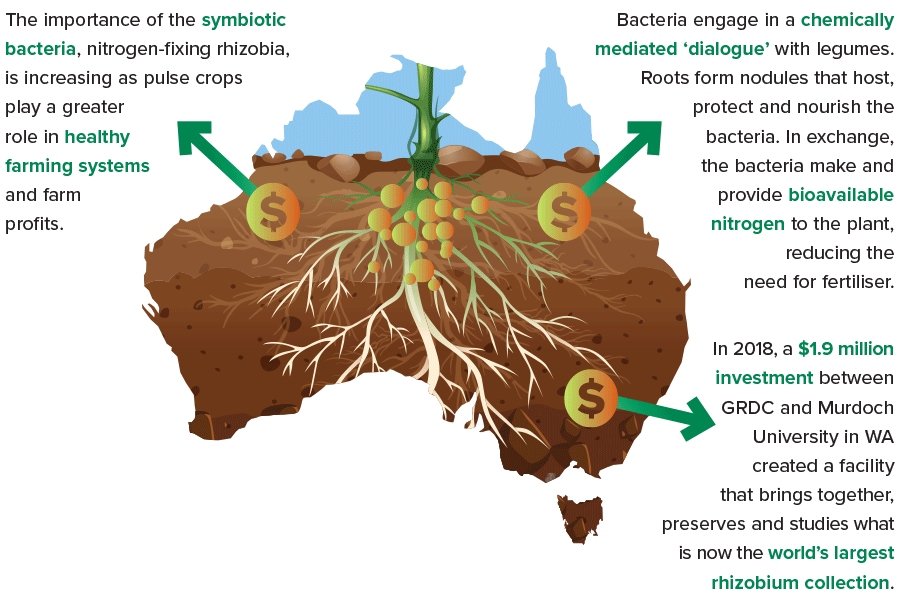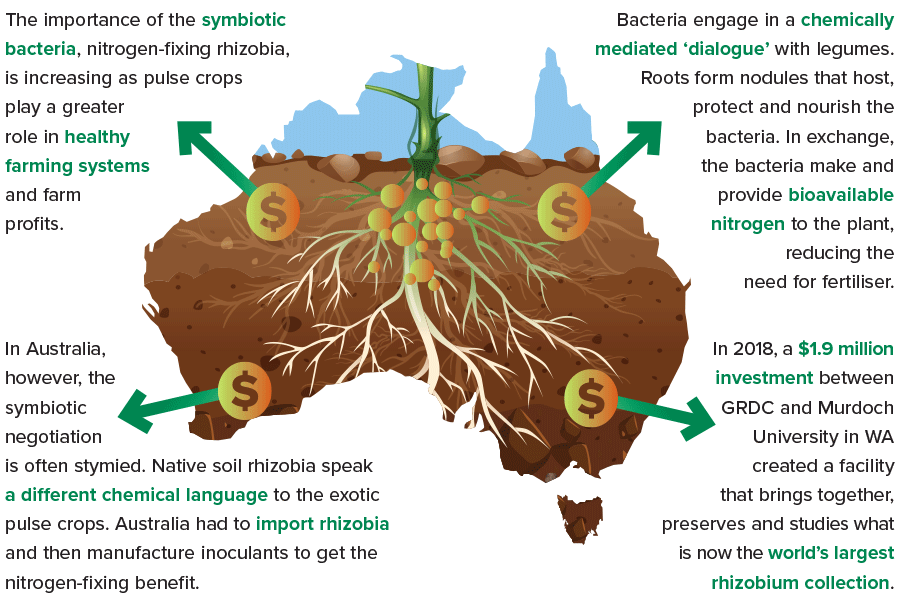
As pulse crops play ever-greater roles supporting the health of farming systems – and farm profits – the need grows stronger to be mindful of a particular class of symbiotic bacteria: the nitrogen-fixing rhizobia.
Early collections
Over 75 years, the nation built numerous rhizobium collections as scientists sought out better-performing inoculant strains that maximise benefits to growers. In attempting to produce this benefit, however, the scientists had to learn how best to mediate in the symbiosis relationship.
This includes understanding which species and strains are the most appropriate for each different legume crop. This ad hoc collection and learning process has now been formally recognised as vital to the grains industry.
Bringing it together
In 2018, a $1.9 million investment between GRDC and Murdoch University in Western Australia laid the foundation for a new facility that brings together, preserves and studies what now amounts to the world’s largest collection of rhizobia.
 Dr Graham O’Hara from Murdoch University’s Legume Rhizobium Sciences. Photo: Murdoch University
Dr Graham O’Hara from Murdoch University’s Legume Rhizobium Sciences. Photo: Murdoch University
The first phase of this endeavour was headed by Dr Graham O’Hara and Dr Jason Terpolilli from Murdoch University’s Legume Rhizobium Sciences, and has now been successfully completed.
Building on this achievement, a second investment has been secured to extend the project for the next four years.
The $2.1 million investment was made through an agreement between GRDC, Murdoch University, and the WA Department of Primary Industries and Regional Development (DPIRD).
Dr Camilla Hill, GRDC manager genetic technologies – pulses, says: “This new investment will ensure the efficient stocktaking of both national and international rhizobia strains housed in the purpose-built facility, supporting sustainable agricultural practices for Australian grain growers.
“It will also support the enhancement and maintenance of an online, interactive catalogue linked to a robust database, streamlining the distribution and accessibility of these valuable strains for the benefit of the agricultural research community, growers and industry stakeholders.”
Save, maintain and build
Dr O’Hara highlights the dire need for the new facility, recounting stories of retiring scientists whose rhizobia collections were in danger of being lost by organisations who were unaware of their significance.
Over time, Murdoch’s Legume Rhizobium Sciences found itself stepping in to rescue these collections, making it the natural site for the new facility, the International Legume Inoculant Genebank (ILIG).
He says that by the end of 2023, the facility held 11,800 rhizobia strains, with capacity for a total of 32,000. This amounts to an amalgamation of 38 previously distinct national and international collections, with the rhizobia sourced from more than 100 countries around the world and going back to the 1950s and earlier (and, therefore, to locations since lost to urbanisation, political turmoil and war).
Contained in this collection are more 90 different rhizobia species from across at least 10 genera. In all, the collection contains bacteria whose symbiotic preferences encompass more than 800 legume hosts.
Samples are maintained in two ways: as lyophilised ampoules (freeze-dried glass capsules) stored in cabinets at room temperature and as frozen samples stored at minus 80°C. A failsafe backup copy of the collection is also stored well away from Murdoch University at a WA DPIRD site in Northam.
GRDC investment in the world’s largest rhizobia collection
A bacterial genebank
The facility, however, does not just store rhizobia. It operates as a functional genebank, providing samples worldwide via an online catalogue.
“We are also setting up an International Rhizobia Network that links up sites around the world that maintain and research important rhizobia collections,” Dr O’Hara says.
Our starting aim is to develop and innovate best practice storage and strain sharing protocols. That will lay the foundation for taking rhizobia research to a new level.
Key to realising new breakthroughs is to sequence vast numbers of genetically diverse rhizobia and systematically decode the genetic decision points that determine whether symbiosis with any particular legume succeeds or fails.
To that end, the facility maintains its own in-house genome sequencing capability, developed with GRDC investment.
The language of symbiosis
Dr O’Hara explains that there are two key steps in symbiosis.
The first is nodulation, in which the bacteria induce the legume roots to form nodules. The second sees the plant feeding carbohydrates to the nodule, which enables the nitrogen fixation process by the rhizobia.
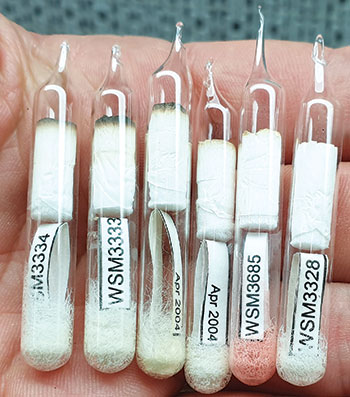 Ampoules containing lyophilised bacteria samples. Photo: Kit Burns
Ampoules containing lyophilised bacteria samples. Photo: Kit Burns
Genome sequencing has discovered that each step requires a core set of 10 to 20 different genes.
It also highlighted that the two steps constitute two distinct barriers to compatibility between host and bacteria that must match for symbiosis to succeed.
For example, Dr O’Hara notes that Australian native rhizobia can sometimes nodulate our exotic legume crops, but the interaction lacks the compatibility needed to initiate nitrogen fixation.
“The host range of different rhizobia strains varies enormously,” he says. “It can be very narrow, such as a preference that is exclusive to Cicer chickpea species, and it extends to incredibly broad, reaching over 100 legumes.”
It is this specificity for particular legumes – along with preferences for soil and climatic conditions – that forms a strong research focus at the centre. This allows the development of new strains for release as inoculants. For example, the centre was recently involved with the release of two new strains. One is highly suited to faba beans, the other to field peas, lentils and vetch.
“Currently, our inoculant development pipeline is limited to blindly screening samples and letting the plant tell us what works and what doesn’t,” Dr O’Hara says.
“With genome sequencing, we can take our phenotyping data to a new level where we can start to predict compatibilities.”
To that end, the genomes of 43 commercial inoculant rhizobia were recently fully sequenced, which brought to light the molecular underpinnings to a broad range of legume specificities.
The nod factor
 Nodulated field-grown chickpea plant. Photo: Kit Burns
Nodulated field-grown chickpea plant. Photo: Kit Burns
As the molecular mechanisms come into focus, the emerging picture resembles a conversation.
The roots release chemicals that signal the bacteria to switch on the nodulation genes.
These genes then produce enzymes that in turn make a chemical signal – called Nod factor – that induces root cell growth and the formation of nodules.
“Nod factor is a molecule that resembles a key designed to fit into a lock,” Dr O’Hara says.
“Different legumes will have different versions of that lock. But different bacteria make keys with different notches – or chemical sidechains.
“It is the differences in the key that mediates the specificity between host and symbiote.”
A bright future
It is these molecular compatibilities that Dr O’Hara’s centre wants to better define.
The aim, ultimately, is to come to know the rhizobia so well – through a marriage of genomic and phenotyping data – that it becomes possible to predict and model the symbiotic interactions.
 Chickpea response to inoculation at a trial in Goomalling, Western Australia, in 2024. Uninoculated chickpea are shown on the left, with a visible contrast to the inoculated plants on the right. Photo: Kit Burns
Chickpea response to inoculation at a trial in Goomalling, Western Australia, in 2024. Uninoculated chickpea are shown on the left, with a visible contrast to the inoculated plants on the right. Photo: Kit Burns
In the immediate short term, the strategy has major pay-offs.
It takes up to 12 years to characterise a rhizobia sample (using existing glasshouse and field study protocols) before it can be released.
Predictive capabilities would allow the development pipeline to function in much more targeted and efficient ways.
It also generates formidable analytical tools. These can interrogate and profile rhizobia populations in soils and nodules in ways that reveal likely impacts on pulse crop productivity.
“Every gain we make readily translates into tools that can support better inoculation management and fertilisation practices,” Dr O’Hara says. “This is as central to agriculture as nitrogen itself.”
Given that something like 15 to 20 new rhizobium species are identified annually, the new facility stands on the brink of something truly monumental: the ability to master biological complexity and become fluent in the language of rhizobia symbiosis.
More information: Graham O’Hara, g.ohara@murdoch.edu.au
Jason Terpolilli, j.terpolilli@murdoch.edu.au

























































