A suitcase-sized diagnostic tool that draws on knowledge from the medical industry is helping crop researchers tackle the pathogens that cause Botrytis grey mould on chickpeas and other pulses.
Biosensors that use functionalised magnetic gold nanoparticles have recently been developed to detect cancer biomarkers with extreme specificity, sensitivity and accuracy. When adapted to the grains industry, these biosensors can accurately diagnose Botrytis-causing fungi in crops in the field in 45 minutes.
Professor Rebecca Ford and her team at Griffith University have partnered with the experts behind the diagnostic tool, also at Griffith University and part of the Queensland Micro- and Nanotechnology Centre, to develop the Botrytis biosensor.
The goal was to adopt this diagnostic approach to detect and quantify plant pathogens. The two Botrytis spp. were used as case studies.
In Australia, Botrytis grey mould on temperate pulse crops is caused mainly by Botrytis cinerea and B. fabae, which infect the crop either separately or as a complex. The disease substantially reduces legume yields during environmentally conducive seasons and can destroy entire paddocks if left unmanaged.
Being able to quickly, accurately and cost-effectively diagnose and quantify the pathogen ahead of symptoms appearing on the plants would provide growers with a huge advantage. If an integrated disease management approach is used, diagnosis and timely management can limit yield and profit losses.
Two-step process
In simple terms, the diagnostic process occurs via two steps.
In step one, target Botrytis spp. DNA are magnetically isolated and purified. In step two, the target DNA is electrochemically detected and quantified.
Figure 1: The two-step process for the electrochemical detection of Botrytis spp. from leaf samples. (A) shows the magnetic isolation and purification of target Botrytis spp. DNA. (B) shows the electrochemical detection and quantification of the adsorbed target single strand DNA.
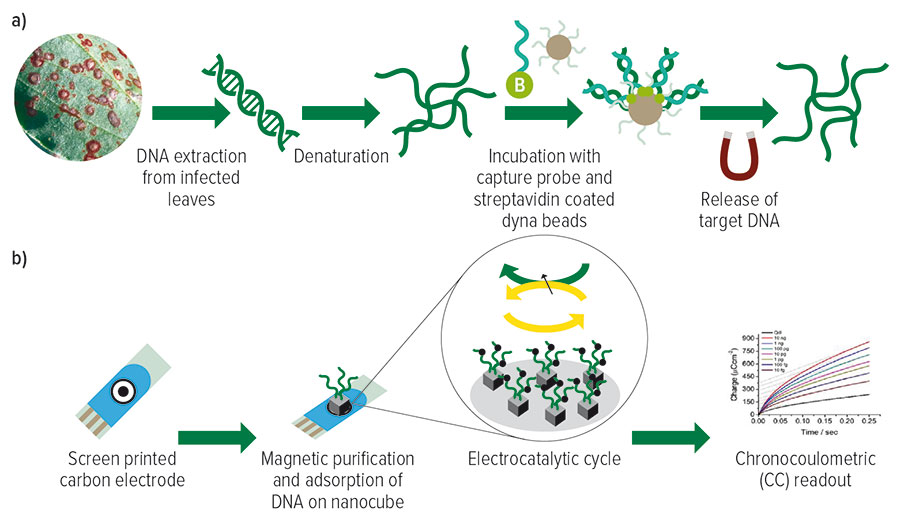
Source: Marzia Bilkiss, PhD thesis, Griffith University
The tool was successfully tested to diagnose B. cinerea in lentil trials at the South Australian Research and Development Institute terrace site, at the Waite campus in Adelaide, and on faba bean leaf samples taken from crops at four field sites in south-eastern South Australia (Millicent, Bool Lagoon, Frances and Mundulla) in October 2020.
Professor Ford says single spores of B. cinerea and B. fabae from symptomatic and asymptomatic leaves were detected. “This demonstrates its ability to detect and quantify the causative organisms prior to the visible appearance of the disease on plants. It is proving to be more sensitive than other published diagnostic methods for both species.”
Not only is it highly sensitive, quantifiable and species-specific to each of B. cinerea or B. fabae, but the toolis alsofast and cost-effective. The process from sample collection to result is about 45 minutes and costs less than $2 per sample.
The tools developed and protocols validated are open for commercialisation.
More information: Rebecca Ford, rebecca.ford@griffith.edu.au

























































