Key points
- Ascochyta blight in chickpeas has become more aggressive over the past five years
- The proportion of isolates collected annually from all major growing regions across Australia where Ascochyta epidemics have occurred that are able to cause severe disease is as high as 70 per cent for PBA HatTrick
- Researchers are targeting pathogen biology and genetics to identify future opportunities to overcome this aggressiveness
Work is underway to limit the increasing aggressiveness of Ascochyta blight in chickpeas.
Ascochyta blight pathogen in chickpea is becoming more aggressive against commonly grown varieties. Caused by the fungus Ascochyta rabiei, it arrived in Australia in the 1970s, most likely on infected seed. Ascochyta has since established across all Australian chickpea growing regions and has rapidly evolved to become highly pathogenic – meaning it has overcome resistance in many varieties.
To develop more-resistant varieties and provide useful advice for growers, we need to understand the mechanisms that enable the pathogen to overcome both genetic resistance and our best management practices.
Resistant isolates on the rise
As part of a six-year GRDC investment (2013–19) researchers from Griffith University and collaborators from NSW Department of Primary Industries, South Australian Research and Development Institute (SARDI) and Agriculture Victoria collected A. rabiei isolates annually from all growing regions where Ascochyta epidemics occurred. Surprisingly, extremely low levels of molecular diversity were detected, irrespective of the source region or chickpea variety.
The ability of these isolates to cause severe disease on a set of resistant lines was measured in plant growth chambers.
Despite the lack of diversity, subsets of these isolates caused severe disease on some of the industry’s most resistant varieties, including PBA HatTrick, Genesis™ 090 and PBA Seamer.
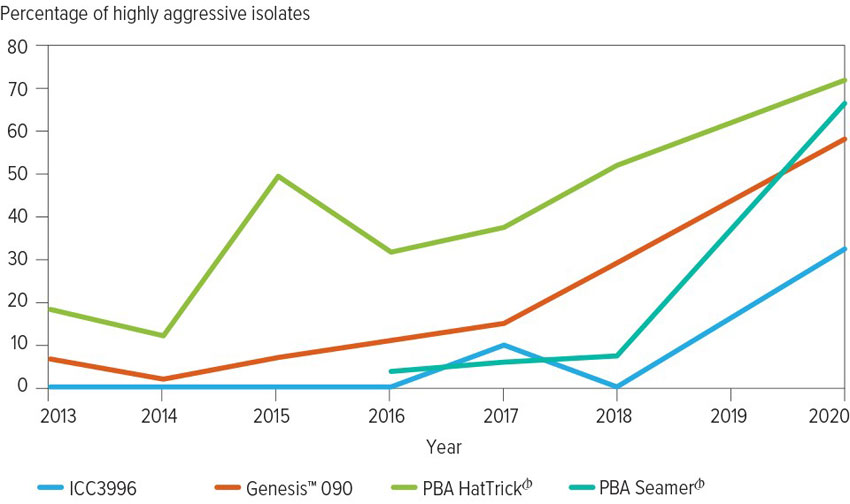
In fact, the proportion of highly aggressive isolates in the background population has increased since 2015 (Figure 1). In 2020, the percentage of isolates sampled from the field that were highly damaging to PBA HatTrick was 70 per cent, 64 per cent were highly damaging to Genesis™ 090 and 62 per cent were highly damaging to PBA Seamer.
Figure 1: In recent years there has been an increase in the frequency of isolates of Ascochyta rabiei that are highly aggressive against common chickpea varieties. Over the past five years the proportion of isolates aggressive against ICC3996 – the breeding line commonly used as a resistance source within the Australian chickpea breeding program – has also increased.
Source: Professor Rebecca Ford
Isolates able to cause severe disease on ICC3996, which is commonly used as a resistance source within the Australian chickpea breeding program, had also increased to 33 per cent in 2020.
The most damaging isolates have been supplied annually to breeders for use in selecting the most resistant lines to bring forward for commercialisation.
The increase in isolate aggressiveness observed in this long-term study has likely occurred through pathogen adaptation, whereby the fungus has evolved to make more highly aggressive isolates in response to selection pressure applied through environmental factors and farming practices.
While external environmental factors cannot be controlled, changes in the pathogen population towards evolving isolates with increased aggressiveness can be reduced by adhering to recommendations for crop rotation and fungicide use.
To understand the ability of the pathogen to evolve to overcome current management recommendations, work at Griffith University is focusing on better understanding the biology, population dynamics and epidemiology of the disease. This is part of the new GRDC investment in collaboration with SARDI, Agriculture Victoria, University of Southern Queensland and Western Australian Department of Primary Industries and Regional Development.
Genetic tools
In their previous work, researchers at Griffith University found that isolates of different aggressiveness levels behave differently on different chickpea cultivars. Some grow and cause severe disease very fast, especially without proper management. The new work aims to identify the genetic basis of isolate aggressiveness. Researchers are looking for molecular sequences that correlate with the ability of an isolate to cause disease on a chickpea variety that contains a particular resistance gene combination.
The aim is to develop a method to ‘pathotype’ an isolate and determine its level of risk to the chickpea industry. A ‘pathotyping tool’ that will quickly and accurately inform growers, pathologists and breeders on the isolate population present at a particular chickpea growing location would help with variety selection and chemical and cultural disease management decisions.
To develop the pathotyping tool, researchers will investigate A. rabiei isolates collected from across all chickpea growing regions to identify genome sequences that are significantly correlated with severe disease incidence. This will also uncover the genetic controls of isolate aggressiveness.
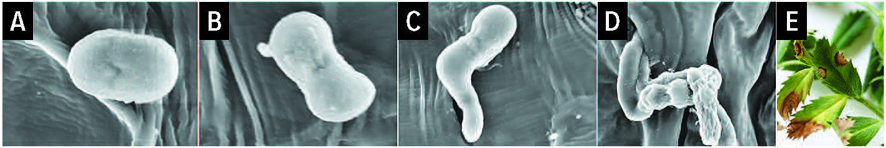
Figure 2: Microscope images of the process of Ascochyta rabiei infection of a chickpea leaf. A) the A. rabiei fungal spore lands on the chickpea leaf; b) the spore germinates (6 to 12 hours); c) the spore grows a germination tube (12 to 16 hrs); d) the germination tube penetrates the chickpea leaf through a stomata opening and colonises the internal tissues (16 to 24 hrs); and e) disease symptoms appear on the chickpea leaves (4 to 7 days).
Source: Yasir Mehmood, Griffith University
This will provide a detailed understanding of the behaviour of A. rabiei and the governing molecular sequences expressed by the pathogen during the infection and chickpea colonisation processes that lead to disease and crop loss (Figure 2). Together with the ‘pathotyping tool’, this will inform the screening for resistance breeding material and disease management packages that are locally relevant.
This research is one of five new four-year research programs invested by GRDC to develop better genetic and management solutions for Ascochyta in chickpeas. The other programs will be led by the University of Adelaide, Agriculture Victoria, CSIRO and the International Center for Agriculture Research in Dry Areas (ICARDA) in Egypt.
More information: Professor Rebecca Ford, 0413 585 439, rebecca.ford@griffith.edu.au

























































