Key message
- Correct identification of viruses in pulse crops is enabling a better understanding of factors that lead to infection, disease and potential yield loss.
New molecular tools are underpinning dramatic improvements in the ability to identify and study viruses in pulses.
Australian losses affecting one or more pulse species have, to date, been attributed to nine different viruses, but in the past decade research has shown much greater diversity than originally thought. There also appears to be large differences within certain virus species in their harmfulness to different host species.
However, symptoms in the field may be the same for different virus species, making effective diagnostic tools a necessity for plant-virus research. Accurate identification is a prerequisite to developing effective control strategies for specific virus-host combinations.
A four-year GRDC investment led by the New South Wales Department of Primary Industries is shedding new light on these microbial pests. The project is using new molecular tools to study pulse viruses in collaboration with Western Australian Department of Primary Industries and Regional Development, Queensland Department of Agriculture and Fisheries, the University of Queensland, Agriculture Victoria and the International Center for Agricultural Research in the Dry Areas.
What’s in a name?
In 2014, widespread virus damage to canola and pulse crops in south-eastern Australia was attributed to beet western yellows virus (BWYV), diagnosed using the antibody assays available at the time. More recently, research using molecular diagnostic tools and whole genome characterisation has revealed that BWYV is actually not present in Australia. Rather, a complex of genetically distinct turnip yellows virus (TuYV) strains was behind the outbreak and preliminary data suggests they may also have biological differences that could affect management and resistance breeding.
Molecular tools have also led to the discovery of two new viruses, also previously misidentified as BWYV – phasey bean mild yellows virus (PBMYV) and faba bean polerovirus 1 (FBPV-1).
Two distinct strains of PBMYV were detected from 11 field host species from across Australia, including seven important grain legume crops. This suggests that its importance in agriculture may have gone unrecognised due to the inability to correctly identify the virus. PBMYV is transmitted by cowpea aphids, not by the green peach aphid – the main vector of TuYV.
FBPV-1 was found in four grain legume species across northern NSW, but as yet there is limited data on its importance in disease outbreaks and the aphid vector remains unknown.
Key differences
We now know that TuYV and PBMYV have quite distinct biological traits, but the genetic diversity within these species may also translate to differences in vectors, pathogenicity and genetic resistances. Preliminary experiments are only just beginning to investigate these differences.
There is still much to learn about the impact of viruses on crops and the factors that will lead to infection, disease or yield loss. Recent research has shown that up to 40 per cent of yield losses in field peas and lentils may result from TuYV infection, despite no visible symptoms.
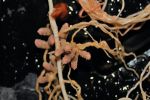
Figure 1: Kaspa field peas (left) are highly susceptible to soybean dwarf virus infection but relatively tolerant, producing very little disease, while PBA HatTrick chickpeas (right) are relatively resistant but highly sensitive once infected.

Source: Dr Benjamin Congdon
Crop susceptibility and sensitivity to a particular virus can vary depending on the environmental conditions, growth stage and even the specific crop variety. For instance, the chickpea variety PBA HatTrick is relatively resistant to soybean dwarf virus infection, but once infected it is highly sensitive, resulting in severe disease that often leads to plant death (Figure 1). In contrast, the field pea Kaspa becomes infected easily but is relatively tolerant, incurring very little yield loss.
More information: Joop van Leur, 0427 928 018, joop.vanleur@dpi.nsw.gov.au