Potential benefits from adopting machine learning technology for grains-based research are being assessed through GRDC investment in nine proof-of-concept projects.
The projects target challenges considered ‘intractable’ by the grains industry and defined as ‘complex’ by scientists.
These are challenges in which crop yield, productivity or resilience are affected not by a single, direct cause, but rather, many variables collude and refer to each other within webs of interaction. These problems are consequently perceived as having no clear causal element for yield fluctuations according to standard analytical methods.
An example is when expression from a network of genes makes concurrent reference to environmental conditions, nutrient levels and pathogen loads. Additionally, gene expression across the entire genome can feel the repercussions from these interactions, which results in variable patterns of yield being associated with that genotype.
GRDC manager of data analytics Dr Jeff Cumpston says current breeding challenges increasingly fall into this ‘too complex’ category, which limits the power of field trials to account for yield variation.
“Breeders are doing all they can to account for the way living organisms respond and adjust to environmental conditions and management practices,” he says. “They have techniques, such as linear-mixed models, to estimate a gene’s contribution to yield. What we are now exploring is whether machine learning provides an opportunity to uncover more complex relationships in the data that went unnoticed with traditional analytical methods.”
Machine learning uses computer algorithms to detect correlations between measurements of crop performance and the outcomes in terms of yield (or grain quality) within the accumulated big data generated by breeding work, pre-breeding research, soil analysis, pathogen diagnostics, climate data and more.
“Machine learning involves detecting patterns within a mix of variables that strongly associate with yield outcomes, with the aim of building a model that can ultimately predict outcomes given real-time measurements,” Dr Cumpston says. “Applied to breeding, for example, this would result in predictions about the best genotypes relative to environmental conditions. This approach is unlimited in the complexity it can handle and can uncover causal interactions that industry currently struggles to detect.”
In a sense, machine learning offers an opportunity to learn from the past by uncovering the complex causal relationships beyond the capability of traditional analysis to detect.
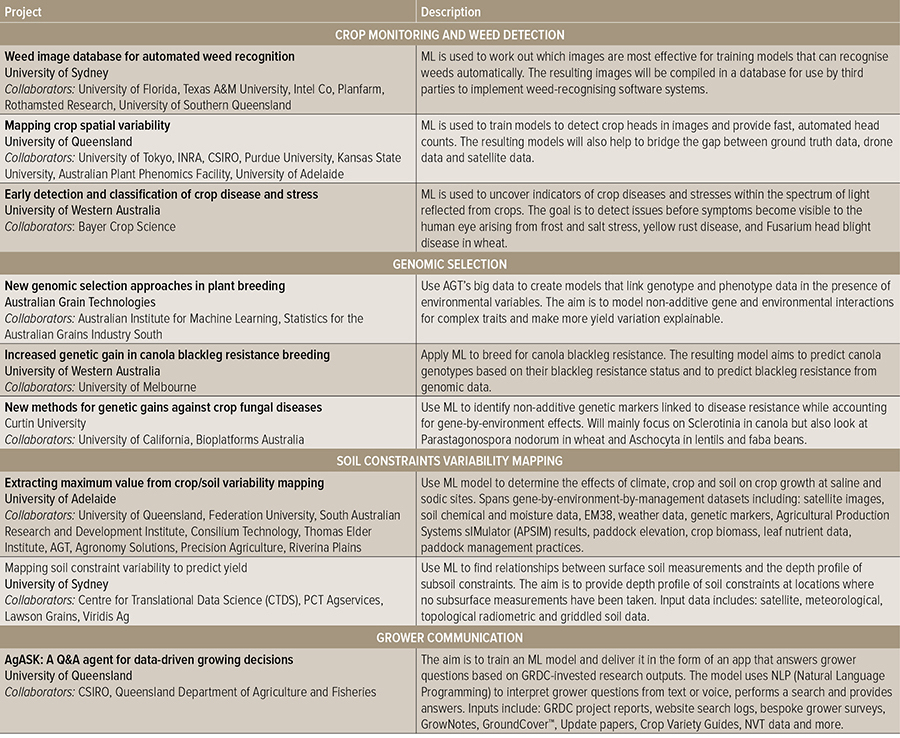
Machine learning capability is under investigation for challenges that fall under four main categories (see Table 1):
- crop monitoring and weed detection;
- genomic selection;
- soil constraint variability mapping; and
- grower communication.
Yield variation
Opportunities around machine learning are especially evident in a genomic selection project headed by Australian Grain Technologies (AGT) in partnership with Statistics for the Australian Grains Industry (South) and the Australian Institute for Machine Learning, both located at the University of Adelaide.
AGT’s Dr Tristan Coram says current methods to associate genotypes and yield involve the use of DNA markers that cover the genome of target crops. Statistical methods and modelling are then used to calculate an effect for each individual marker that corresponds to its contribution to yield. These are then added to give an estimate of a genotype’s value to the breeding program.
Not all gene effects on yield can be accounted by simply adding them together. In addition to these ‘additive’ effects are more complex interactions, called ‘non-additive’.
“Current methods cannot fully account for all the yield variation we detect in field trials,” Dr Coram says. “We know that a lot of this unexplained yield variation arises from gene-by-gene and gene-by-environment interactions and they give rise to complex ‘non-additive’ impacts on yields.”
The project has two modules depending on whether or not information about the environment is included. The simpler restricts the modelling to finding hidden associations between yield and the genome-spanning DNA markers already in use at AGT. The second will train the yield-predicting model to take environmental conditions into account, including spatial soil variability data (nutrient profile, electroconductivity and pathogen load) and weather data.
“The goal is to integrate the environmental data with the genotype data and identify the key parameters affecting yield,” Dr Coram says. “That places us in a position to build a model that can predict yield of breeding material in a new environment without ever testing it there.”
Ultimately, the two-year project is looking to improve on existing statistical modelling and determine whether there is a path forward to increase genetic gain for yield into the future.
“We’re generating good results, but there is still lots of optimisation needed around identifying the parameters that affect yield,” Dr Coram says. “Machine learning is an interesting new technology and we can see the potential, so the opportunity to do this scoping work has been exciting.”
Table 1: The nine projects supported for two years through the GRDC’s Machine Learning (ML) initiative.
More information: Jeff Cumpston, jeff.cumpston@grdc.com.au; Tristan Coram, tristan.coram@agtbreeding.com.au

























































