Ascochyta blight is a continual threat to chickpea production in Australia but a forensic approach is set to type both pathogen and chickpea genomes and improve the odds of success in the battle against the disease.
If you know your enemy and know yourself you are much better prepared for any battle – and it is this approach which is being taken to better inform management of Ascochyta blight in chickpeas.
Internationally, studies on pathogen diversity have been conducted on a country-by-country basis, using different chickpea genotypes and without identifying specific resistance genes linked to particular pathotypes.
Unlike cereal pathogens, the interaction between chickpeas and Ascochyta blight (Ascochyta rabiei) suffers from a lack of known resistance genes or agreed differentials, as well as sufficient data on factors driving population changes in the pathogen.
To address this, a project led by the International Center for Agricultural Research in the Dry Areas (ICARDA) and the Centre for Crop Disease and Management (CCDM) atCurtin University aims to develop a set of chickpea lines with known markers for each resistant gene in the chickpea host plants and identify corresponding avirulence or virulence genes in the pathogen. This forms part of GRD’s five pronged approach ‘Towards effective genetic and sustainable management of Ascochyta blight of chickpea’. It involves creating segregating fungal populations of the pathogen to aid in the discovery of (a)virulence genes and determine the pathotype structure for the species.
Ultimately, this project will make novel Ascochyta blight-resistant genes accessible to Australian and overseas chickpea breeders, leading to improved Ascochyta blight resistance in future varieties.
By understanding the pathotype structure and (a)virulence genes discovered, chickpea growers worldwide will be empowered to enhance their production and profitability. This endeavour holds the promise of bolstering chickpea cultivation and ensuring food security not only in Australia but also on a global scale.
Fungal fingerprinting
The CCDM team obtained 39 A. rabiei isolates from international collaborators and generated high-quality chromosome level genome assemblies for 13 of these isolates. Comparing these international isolate genomes to Australian ones revealed that Australian isolates have much less sequence diversity (Figure 1).
Figure 1. Whole genome relationship of 12 Australian and 13 international Ascochyta blight isolates sequenced to date. The 12 Australian isolates all cluster together and have a narrow genetic diversity when compared to the international isolates.
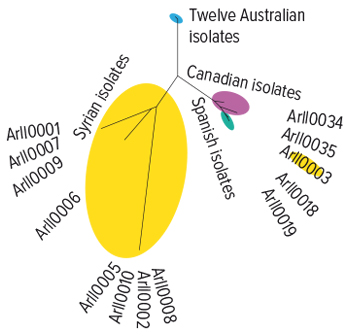
Source: Robert Lee, CCDM
The low level of genomic diversity in Australian A. rabiei isolates is most likelydue the presence of only one mating type in Australia and thus sexual recombination does not occur in Australian A. rabiei isolates.
Hence, a team led by Professor Rebecca Ford at Griffith University (part of GRDC’s five-pronged Ascochyta blight investment) each year is screening A. rabiei isolates collected in Australia for their mating type to ensure the second mating type remains absent.
At Curtin University’s biological containment facility, the team generated four fungal mapping populations to identify key genes in the pathogen’s genome associated with causing disease. This required the development of a robust phenotyping process in the containment facility for Ascochyta blight resistance (Figure 2).

Figure 2. Chickpea seedling Ascochyta blight assay in the misting hoods at Curtin University. Photo: Robert Lee, CCDM
This method has been optimised to evaluate the response of the international isolates on 15 ICARDA differential chickpea lines as well as the Australian chickpea differential set used in other projects within the GRDC’s Ascochyta blight five-pronged investment. Currently the team has phenotyped the disease response of 83 offspring of the first fungal population.
Global chickpea germplasm generation
A global consortium was formed by the project involving several countries (Australia, Tunisia, Turkey, Lebanon, Ethiopia, India, and Morocco) to tackle Ascochyta blight in chickpeas. The scientists in these countries collected more than 150 isolates of the pathogen from different regions and tested their pathogenicity on various chickpea varieties under controlled conditions (Figure 3).

Figure 3: Assessment of the pathogenicity of isolates collected from various locations. The evaluation was carried out in both a growth room and greenhouses to maintain controlled conditions and prevent contaminations between isolates. Photos: Aladdin Hamwieh, ICARDA, and Canan Can, Gaziantep University.
A range of responses was observed, with isolates showing low to high pathogenicity across all countries. The DNA of these isolates was sent to Australia for sequencing to gain a better understanding of the global diversity in the pathogen population. Additionally, the project created a collection of chickpea genotypes called the global Ascochyta blight resistant subset collection (GABRSC), which included various Ascochyta blight-resistant genes from different sources.
Seeds from this collection were shared among consortium members and tested in multiple locations in the participating countries for two years (2022 and 2023). The results revealed that certain genotypes (S0110227, S160454, S0110088 and S0110028) consistently showed resistance to Ascochyta blight irrespective of the country location and isolates used (Figure 4).

Figure 4: Field evaluation of GABRSC performance in during the 2022 growing season in the field (A) in Béja station (north-west of Tunisia) and (B) in Kafarshkhna station (West Lebanon). The experiments were laid out in an Alpha Lattice design, with two replications, and conducted under natural infection. Photos: Tawffiq Istanbuli, ICARDA and Mariem Buhadida, INRAT, and Canan Can, Gaziantep University.
These chickpea genotypes were developed at ICARDA through the strategic combination of resistant parent plants over the past four decades, presenting a promising approach to creating sustainable resistance against Ascochyta blight in chickpeas. The GABRSC population is currently under quarantine with the Australian Grains Genebank and material will be disseminated to other projects within GRDC’s Ascochyta blight five-pronged investment for evaluation with Australian isolates.
At ICARDA, more than 300 chickpea near-isogenic lines (NILs) were established. NILs are a valuable and fascinating tool in the field of genetics, designed to address complex traits such as Ascochyta blight disease in chickpeas. The chickpea NILs are a pair of lines that are genetically similar but only differ in a small region of the genome. In our case, this region contains a single Ascochyta blight resistance gene in one line and not the other.
This unique structure allows researchers to isolate and study the effects of individual Ascochyta blight-resistant genes, making it easier to understand the genetic basis of characteristics of each resistant gene.
By comparing NILs to the original susceptible chickpea line ILL263, scientists can pinpoint the precise genes responsible for Ascochyta blight resistance, and develop markers to follow the different resistance genes in breeding programs. This will lead to significant advancements in chickpea crop improvement and ultimately contribute to global food security and sustainable agriculture.
More information: Dr Aladdin Hamwieh, [email protected], Dr Lars Kamphuis, [email protected]

























































