Key points
- Pulse crop improvement using genomic tools is essential for accelerating trait genetic gains
- Technology needs to be standardised to link germplasm development to commercial variety delivery for the Australian pulse industry
- A custom-made multispecies SNP chip has been developed. It will improve efficiencies in understanding the genetic basis of important pulse traits and accelerating their incorporation into the development of new varieties with improved yield
Efficient delivery of valuable traits for pulses requires the development pipeline to be connected and accelerated, from germplasm identification by pre-breeders through to commercial breeding.
However, to date there has been no standard technology available for pulses, which makes it difficult to connect research findings across different projects and to link pre-breeding outcomes with breeding. This is a major gap that slows the adoption of research outcomes by breeders and consequently development of new varieties.
Agriculture Victoria Research (AVR), with investment from GRDC, is developing a unified DNA-typing platform for chickpea, lentil, field pea, faba bean and lupin. To achieve this, the project is using a proven genotyping technology known as Infinium SNP chip arrays.
A SNP (pronounced 'snip') is a DNA marker designed to detect differences of one nucleotide (one letter in the genetic code) at a specific region of the genome. A SNP chip deploys thousands of such markers to detect genetic differences across the entire genome. The chip can then be used to rapidly screen, type and compare genetic diversity within a population. This includes diversity linked to valuable traits.
This technology has been used for human, animal and crop genotyping (for example, wheat and barley) for more than a decade. A recent advancement enables genotyping assays for multiple crop species to be placed on the same SNP chip, which reduces set-up costs. This has enabled AVR to develop a cost-effective multispecies SNP chip for pulses.
To ensure the set of markers used in the SNP chip is the most informative for both pre-breeding and breeding applications, AVR consulted with breeders and researchers as well as the Australian Grains Genebank (AGG) in Horsham. This included leveraging the foundational work of CSIRO to develop the Lupin Breeders' Toolkit.
These consultations identified germplasm that captured worldwide genetic diversity. After initially genotyping about 5000 pulse lines, a subset of 250 lines per species were selected that captured the maximum diversity. Their genomes were then sequenced. Mathematical approaches were used on the sequence to identify a subset of the most informative genetic variations for use as markers in the SNP chip. This was done for each pulse species.
The custom-made SNP chip offers unprecedented genetic resolution to more rapidly dissect and understand the genetic basis of pulse traits.
Statistics overlaid on genetics
The SNP chip content was selected using advanced statistical methods (called imputation) to ensure that the information produced by the SNP chip array captures most of the genetic variation present in the tested pulse germplasm.
This ensures the SNP chip can optimise the exploitation of genetic diversity to the benefit of the pulse industry. At the pre-breeding end of the development pipeline, the SNP chip data can be used to correlate valuable traits that were detected in field trials with the inheritance of specific SNP markers. This means the new pipeline has a readily deployed tool to understand the genetic basis of traits.
Once the association between trait and genetics is established, the SNP chip technology allows breeders to rapidly and routinely select for that trait during breeding. This new capacity then facilitates the development of pulse varieties with improved yield, disease resistance, better adaption to local growing conditions and resilient to future climates.
Seamless integration
This project will also ensure that data generated using the new SNP chip platform can be used to seamlessly link research outcomes across pre-breeding projects, as well as facilitating the adoption of research outcomes in breeding for accelerated variety development.
This is being achieved using a new web-based data visualisation tool (called Pretzel) developed in another GRDC project. This tool allows researchers to immediately ask whether a trait locus (a specific, fixed position on a chromosome where a particular gene or genetic marker is located) identified in their study is novel or if it has been previously described. It also allows them to question a range of datasets to identify candidate genes underlying the desirable trait (see Figure 1).
Figure 1: Pretzel visualisation of datasets generated across time. Comparison of a genetic map generated in 2006 (a) with positions of quantitative trait loci (coloured boxes) and gene models (orange lines) generated in 2018 (b).
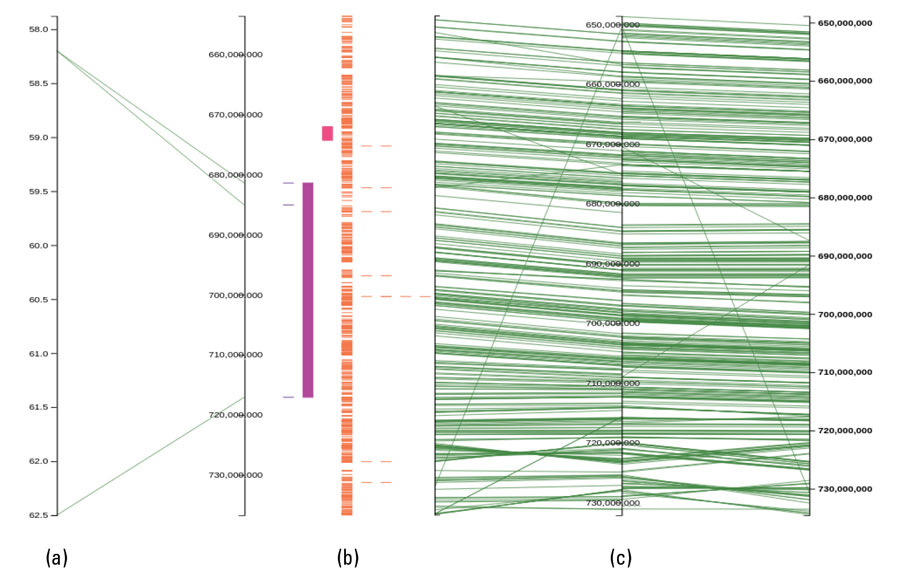
Source: AVR.
In the near future, one of the project's anticipated outcomes is the ability for researchers and breeders to select the best suited germplasm from the AGG for a particular study based on information from the new SNP chip. This will allow trait variation present in AGG material to underpin the development of future pulse varieties with higher yield potential, superior nutritional quality and increased stress tolerance to meet current and future production challenges.
More information: Dr Sukhjiwan Kaur, 03 9032 7000, sukhjiwan.kaur@agriculture.vic.gov.au

























































