Key points
- Understanding the genetic basis of crop phenology is key to better matching crop varieties to target environments
- A huge collective, multidiscipline effort is underway to provide growers access to accurate predictions of wheat and barley cultivar flowering dates from a broad range of sowing dates
Phenology is the study of cyclical events in an organism’s life cycle. For plant breeders it has historically been a way to breed crops that are better-matched to environments. With recent scientific advances by integrated multidisciplinary research teams, GRDC is soon to deliver improved phenology information to help growers optimise crop productivity.
Understanding the genetic basis for variation in phenology can inform crop breeding strategies and contribute to prediction of yield risks, such as drought, frost or heat, and thereby improve crop management.
An understanding of phenology for different crop varieties enables specific variety and sowing time selection to ensure flowering occurs at the optimum time for a particular location to maximise productivity (see Figure 1).
Figure 1: The different growth stages, optimum sowing and flowering windows and the dominant environmental signals that influence growth and development in each stage in wheat.
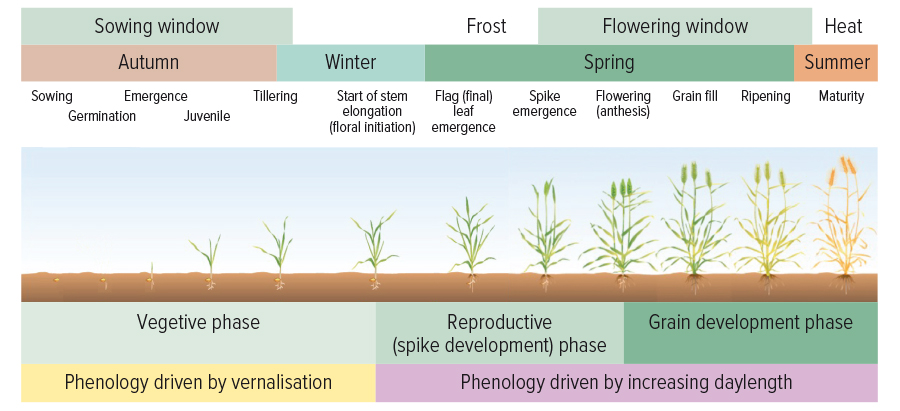
Source: GRDC
Phenology pipeline
For plant breeders and growers, there are key components involved in understanding phenology to improve plant productivity.
Since phenology can be under the control of multiple genes, the genetics that control phenology need to be investigated first, and this information can then be used in models to determine the influence of the environment on various genetic combinations. From this, crop variety and sowing time can be selected to better match specific locations to optimise yield.
Described as the ‘Phenology Pipeline’ (see Figure 2), GRDC has invested in this research for many years and its portfolio of crops are at different stages of maturity within the pipeline. Wheat and barley are the most advanced whilst canola is part way, and oats, chickpeas and lentils have only just commenced.
Figure 2: GRDC’s phenology pipeline showing the progress of various crop species along the pipeline.
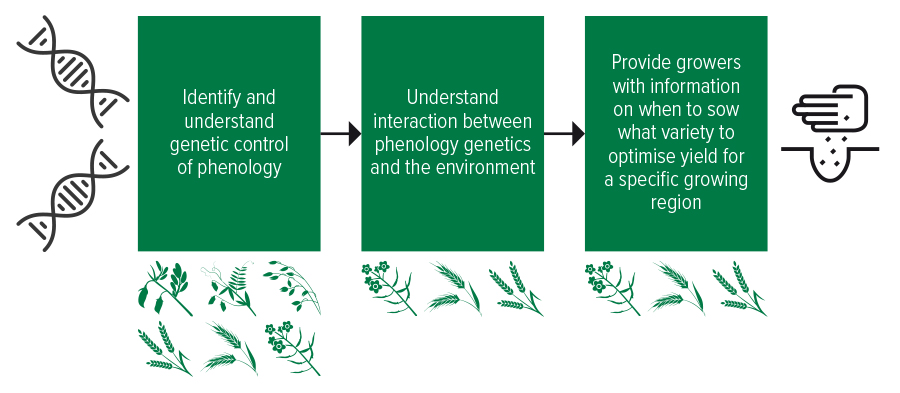
Source: GRDC
Gene discovery

Working to discover new phenology genes to better match Australian crop varieties to environments. Members of the CSIRO team that form part of the seamless pipeline delivering for Australian growers; Jessica Hyles (right), CSIRO Team Leader and University of Sydney PhD student and Tina Rathjen CSIRO Research Projects Officer. Image: CSIRO
For 20 years, the genetic basis of plant phenology has dominated the research of a broad team at CSIRO Agriculture and Food. With GRDC investment, it has been dissecting the genetic control of phenology at the start of the phenology pipeline.
“Phenology is a major driver of adaptation for plants and – most crucially for grain plants – is a driver of flowering time and, therefore, yield,” says Dr Ben Trevaskis, speaking on behalf of colleagues at CSIRO.
“It is controlled by many genes with major and minor influence and these respond to environmental stimuli.
“Two important stimuli are daylength, or photoperiod – the cycle of light and dark periods – and temperature, particularly a period of cold required by some plants to trigger flowering, which is known as vernalisation,” Dr Trevaskis says.
Crop plants can be described on a scale as having varying speeds of development from spring to winter habits dependent on their responses to these environmental stimuli.
Of particular importance is resolving the basis of this genetic control and also gene-environment interactions to understand and predict the flowering behaviour of diverse crop varieties at a range of locations.
“A decade ago, we were using molecular biology to study individual genes, one at a time, which was labour-intensive and slow. But the science has developed significantly, and we now use powerful, high-throughput genomic sequencing to provide detailed insights into the genetic makeup of plants,” Dr Trevaskis says.
“Also incredibly valuable are the breeding pedigrees of plant varieties developed over many years by plant breeders."
“We can use a big data approach to interrogate this information and match it to the genetic sequencing information to create gene databases for plant breeders, which can then inform breeding programs. For example, it can identify appropriate parent plants to cross to obtain a desired genetic combination in the offspring.
“We are also exploring the application of machine learning to trait prediction, using flowering behaviour as a test system.
“This research aims to integrate multi-level genomic data with detailed environment characterisation to resolve gene-environment interactions and predict traits under field conditions. It could significantly accelerate scientific discovery.”
Researchers at CSIRO are actively exploring this use of machine learning to accelerate breeding in the CSIRO Future Science Platform for Machine Learning and Artificial Intelligence.
“Collaboration, with the seamless integration of components along the phenology pipeline, is key to the success of delivering these traits to Australian growers,” Dr Trevaskis says.
“In the genetic area, we have long-term collaborations with other Australian grains industry researchers located with the ARC Centre of Excellence for Plant Success in Nature and Agriculture (University of Queensland), Queensland Alliance for Agriculture and Food Innovation (QAAFI), La Trobe University, University of Tasmania, University of Adelaide, and NSW Department of Primary Industries (DPI), as well as international researchers at Oregon State University (US), the Chinese Academy of Science, Department of Botany, the International Maize and Wheat Improvement Center in Mexico, University of California, Davis, in the US, John Innes Centre in the UK and IPK Gatersleben in Germany.
“But equally important is our collaboration with Australian-based farming systems researchers, where we learn about the environmental and agronomic fit for the genetics we study.”
It is through the tight connections along the pipeline, with farming systems research teams such as those led by Dr John Kirkegaard at CSIRO and Associate Professor James Hunt at La Trobe University, that Dr Trevaskis gleans information on the agronomic significance and business cases of phenological traits for Australian cropping environments on which to focus his team’s research.
National Phenology Initiative
Associate Professor James Hunt leads the GRDC National Phenology Initiative (NPI), a cross-functional team of more than 22 agronomists, crop physiologists, geneticists, modellers, data scientists, bio-informaticians and web programmers from La Trobe University, CSIRO, Plant & Food Research NZ, the Western Australian Department of Primary Industries and Regional Development, the South Australian Research and Development Institute, NSW DPI and Statistics for the Australian Grains Industry West. The team is in the process of building a phenology platform to support Australian wheat and barley industries into the future.
“Growers need to know about the phenology of new crop varieties to determine optimum sowing time for their environment,” Dr Hunt says. “But there has not been a recognised industry-wide scale for plant development, consistent terminology or a readily available tool to determine sowing dates.”
As part of the project, the team has been developing an industry-wide crop development scale to be used by researchers and agronomists, and working with Australian plant breeders to standardise phenological wheat maturity descriptions (see Table 1).
Table 1: Description of industry-wide endorsed maturity classes for wheat, which will be updated annually.
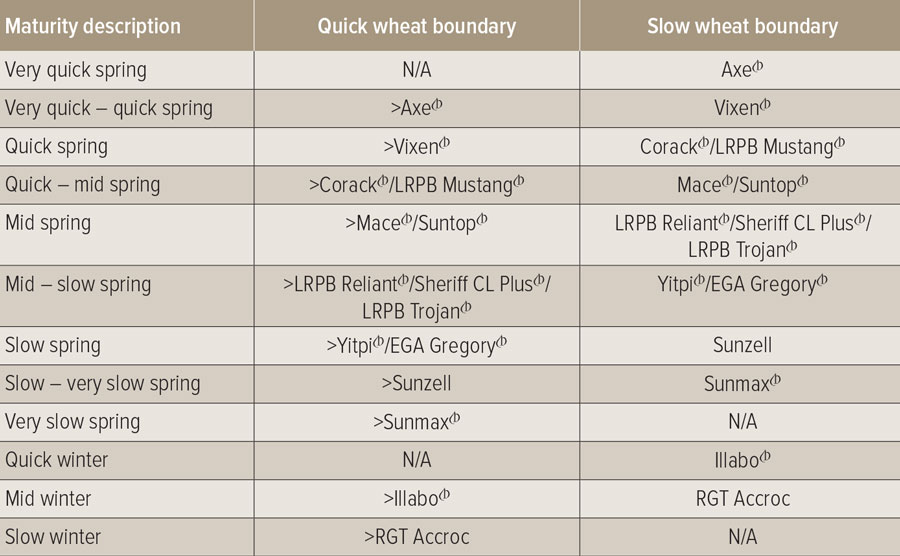
Source: Australian Crop Breeders
“The NPI’s core work is building on the plant modelling power of the next-generation Agricultural Production Systems sIMulator (APSIM) – a massive piece of research infrastructure initiated by Agricultural Production Systems Research Unit in the early 1990s,” Dr Hunt says.
“A total of 64 wheat and 32 barley genotypes, selected for their diverse phenology, are being genotyped and phenotyped under different controlled conditions to parameterise the model.”
The same genotypes are also being grown and phenotyped at five field sites in WA, SA, NSW and Victoria, and across eight sowing times to validate the model.
This can provide information about yield losses related to the impact of frost and heat, which are still difficult to quantify for a given environment. The NPI provides a foundational piece of the puzzle in helping optimise trade-offs with frost risk, heat risk and soil moisture deficit in spring.
“The activities are integrated across the phenology pipeline and are fundamental to its success, as we rely on the genetic information from Ben Trevaskis’ team to provide insights into the biological significance of the genes.”
“It will then be straightforward to adapt the model for other crop species to provide similar information for growers’ sowing decisions.”
More information: Associate Professor James Hunt, 0428 636 391, j.hunt@latrobe.edu.au; Dr Ben Trevaskis, 02 6246 5045, ben.trevaskis@csiro.au

























































