Mass screening of chickpea lines is being combined with genomic resistance identification to improve the odds for chickpea’s fight against Ascochyta blight.
Enhanced, durable resistance to Ascochyta blight will aid more-reliable, lower-cost production for chickpeas across existing production areas while also promoting possibilities of expansion.
However, genetic resistance to chickpea Ascochyta blight globally appears to be limited, despite significant research efforts. It is the primary disease constraint for chickpeas in Australia.
The most-resistant variety (Genesis 090) is rated moderately susceptible with up to 64 per cent yield loss, with susceptible varieties such as PBA Striker suffering up to 96 per cent loss.
Since 2020, GRDC has invested in a five-pronged program for Ascochyta blight with partners across Australia and internationally. Program 3 ‘Towards effective genetic and sustainable management of Asochyta blight in chickpea’ is led by the South Australia Research and Development Institute (SARDI) by Dr Janine Croser and a multidisciplinary team incorporating researchers from SARDI, the University of Adelaide and Agriculture Victoria Research (AVR).
This partnership galvanises a holistic scientific approach to improve chickpea Ascochyta blight resistance. Field-relevant pot-based phenotyping capabilities at SARDI assess well adapted as well as poorly adapted germplasm such as landrace and wild relatives that would struggle to grow under field conditions. Field disease nursery screening at AVR, led by Dr Joshua Fanning, provide scalable, high-throughput grower-relevant disease response assessments.
The high correlation between the two phenotyping approaches means the data collected can be combined in the same analysis increasing statistical power to understand Ascochyta blight’s genetic response. Similarly, trait dissection at SARDI and The University of Adelaide identifies functional genes that underlie Ascochyta blight disease resistance. Genomic prediction and speed breeding work led by Dr Sukhjiwan Kaur’s team at AVR unravels the genetic basis of the disease response and identifies favourable alleles for resistance that are rapidly combined into seed stocks, ready for deployment in breeding. Underpinning these efforts is the multispecies pulse DNA chip, recently developed by Dr Kaur’s team. This chip more seamlessly connects pre-breeding research outputs to breeding outcomes.
Ongoing breeding efforts for Ascochyta blight resistance in chickpea, under the mantle of Chickpea Breeding Australia (CBA), will be fortified with outputs from these new pre-breeding efforts. Resistance levels are building as CBA pipeline material recorded no yield loss last season when exposed to Ascochyta blight infection. This effort will be constantly required as the pathogen is highly mutable, which means it keeps changing, creating an ‘arms race’ between researchers/breeders and the disease.
Mass screening
The project has assessed Ascochyta blight resistance in 2465 different chickpea lines in the field and 1600 lines in a pot-based adult plant screening facility at SARDI.
These lines are as diverse as possible and include CBA breeding lines, international breeding program lines and wild relatives supplied by the International Center for Agricultural Research in the Dry Areas (ICARDA Food Legume Improvement Program-FLIP) and the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), landrace lines (Vavilov) and wild relative (Cicer reticulatum and C. echinospermum) accessions. The international and wild relative germplasm seed were supplied by Dr Sally Norton’s team at the Australian Grains Genebank.
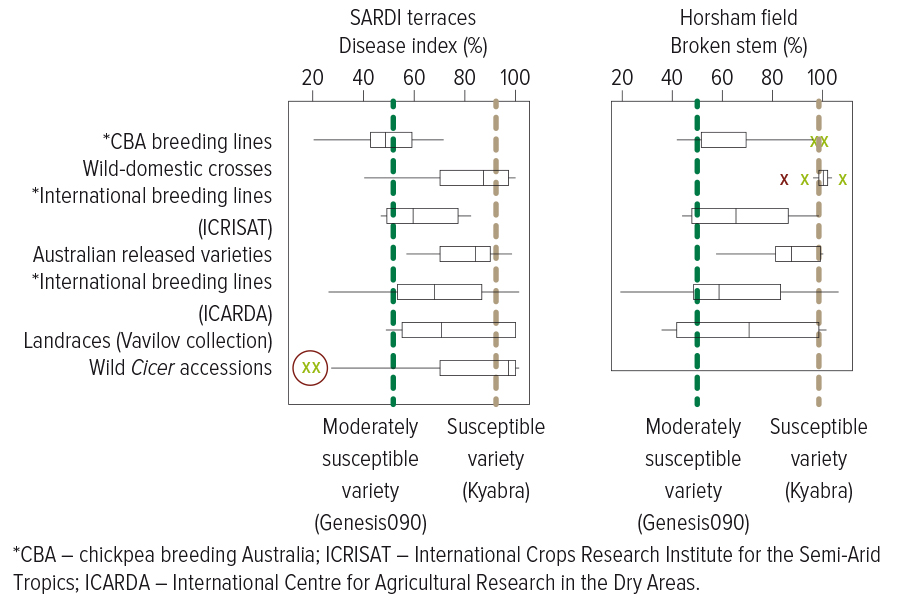
Disease assessment has been undertaken over three years and in two environments. The first, the SARDI ‘Terraces’, is a bird-net protected facility on the Waite Campus in Adelaide that provides a field-relevant, pot-based phenotyping platform in outdoor conditions. These pot-based methods enable adult plant assessment of poorly adapted germplasm.
Plants in the Terraces are assessed for resistance to Ascochyta blight based on four measures: per cent main stems broken, per cent stems with lesions, per cent side branches with disease and per cent leaves with disease to develop a disease index rating. This comprehensive rating provides the data required to help understand the biological mechanism of resistance.
The second environment is the Horsham field disease nursery, undertaken in partnership with Dr Josh Fanning and his team at AVR. Field assessment is based on per cent stem breakage. This project has demonstrated an excellent correlation between the Terrace and field environment, providing evidence of the field-relevance of the pot-based screening methodology. It has also shown the value and scalability of field-based screening for high-throughput disease screening.
Figure 1: Comparative performance of chickpea lines to Ascochyta blight in the SARDI terraces (left) and in the field at Horsham as compared with moderately susceptible (Genesis 090) and susceptible (Kyabra) commercial lines. The red circled wild Cicer accessions show additional genetic potential within wild relatives.
SARDI
By combining the data from these two approaches 86 promising lines were identified. Seed from promising lines has been harvested and provided to CBA to ensure breeding efforts can rapidly exploit novel resistance sources.
Genomic work
Concurrently, Dr Judith Atieno (SARDI) and Dr Ute Baumann (The University of Adelaide) have been working to identify genomic regions associated with resistance, using a method called Genome Wide Association Studies. This is a statistical method linking phenotyping information from SARDI with genotyping information from AVR.
This approach shows novel areas of the genome that are associated with resistance and has highlighted that several genes might need to be combined into a single genotype to develop durable resistance. The novel genomic areas have been targeted using sequence capture and this information will lead to a better understanding of the genetics of Ascochyta blight resistance.
To enhance the genetic analysis, SARDI has assembled the genome of Australian variety PBA HatTrick. The PBA HatTrick genome and further information is now publicly accessible.
In parallel, the AVR team focused on implementing a genomic selection approach to identify and combine through breeding both the major genes as well as minor genes that are responsible for Ascochyta blight resistance. This approach mitigates the risk of breakdown of the major disease resistance genes which occurs frequently due to the pathogen evolving over time.
Their research found that the newly identified large-effect resistance sources explained about one-third of the genetic variance for Ascochyta blight resistance, while the remaining two-thirds was explained by small-effect genes that are mostly already present in domesticated chickpea germplasm.
AVR has combined genomic selection with speed breeding approaches to develop seed stocks that are enriched with both major and minor gene resistance. The speed-breeding facility at AVR fast-tracks the development of fixed lines by reducing the generation cycle time of plants to eight to nine weeks, in comparison to more than six months in the field. A subset of these improved seed stocks is under validation in the Horsham nursery as well as SARDI Terraces. Once validated, these stocks will be delivered to CBA.
The AVR Speed Breeding team. From left to right: Shahin Yazdifar, Laura Spooner, Melanie Scatchard, Elsa Gurban and Amardeep Kaur. Photo: Adam Dimech
In the future
The pot-based phenotyping has identified wild relatives of domestic chickpeas with higher resistance to Asochyta compared to the cultivated species. Importantly, novel resistance has been found in the close relative Cicer reticulatum, which has been crossed at SARDI to domestic germplasm for incorporation in future breeding and research activities.
Utilising a genomic selection approach to further exploit current variation for Ascochyta blight resistance within cultivated germplasm will be important, as well as simultaneously and rapidly introducing newly identified major sources of resistance into new varieties. The molecular tools being developed by AVR will greatly assist breeders to achieve this.
The research teams will continue to seek opportunities to enhance the PBA HatTrick genome assembly to take it to a level of accuracy and relevance for Australian breeding germplasm beyond what is currently available.
More information: Dr Janine Croser, [email protected], 0448 990 281

























































