Identification of phenology genes and acquisition of more-diverse germplasm will expand the regions where chickpeas can be grown in Australia.
Phenology, or the timing of key events in a plant’s life cycle, is a crucial trait that breeders harness when improving crop adaptation and performance. As climates become more unpredictable, phenology is becoming an increasingly important consideration in developing new varieties that are productive across a wider range of environments and climatic variables.
As such, understanding what controls phenology and being able to manipulate it more effectively through breeding are two important components in a far-sighted strategy for climate adaptation.
Compared to other crops, relatively little is known about the genes that control phenology in chickpeas. Discovering these genetic factors and how they intersect to determine phenology in different environments is one main goal of a GRDC-invested project underway at the University of Tasmania in partnership with the South Australian Research and Development Institute and CSIRO.
This knowledge will be an important asset in breeding programs for several reasons. It will provide breeders with new information and, when combined with genetic markers necessary to track specific gene variants, will enable selection for combinations of genes that confer diverse phenologies. It will also enable the development of genetically defined lines that help to assess the value of different phenologies for different locations and management options.
The Australian chickpea industry was founded on a relatively narrow subset of the genetic diversity available for the species globally. This means that breeders had limited access to the variation needed for the development of improved and diversified varieties. To address this, the project is examining a much-wider range of germplasm acquired from around the world that represents new diversity for phenology and related traits.
Identifying phenology genes
In many grain crops, there is good basic knowledge of genes that control flowering and progression through the reproductive phase. However, in chickpeas only one major gene controlling flowering time has been identified so far - early flowering locus 1 (efl1). This gene pushes phenology earlier, promotes pod filling and hastens maturity, and is important for short-season environments globally, where escaping terminal drought is a key problem.
In addition to efl1, the importance of at least five other genomic regions has been reported. To examine several of these in detail, researchers have generated populations from crosses between early and late lines. These populations have been grown in controlled-environment facilities at the University of Tasmania and assessed for variation in phenology, including the time of flowering, flower abortion and pod development.
The genetic analysis of these populations is currently underway and will allow researchers to better define several other major gene variants, including those that delay flowering. The researchers have already identified a second gene which provides a novel, and potentially contrasting, source of earliness to efl1.
One of the benefits of this detailed study is that, as different genetic means to achieving earlier or later phenology are identified, different genes are expected to have a different spectrum of effects on other traits such as growth habit, and also respond differently to environment. This will provide opportunities to expand the environmental regions of production for chickpeas.
The identification of specific genetic markers is another intended output from the project that can be used to follow the genes through the breeding process, and select specific combinations that may be desirable in a certain context. This is particularly important in situations where there may be alternative combinations with similar effects that need to be distinguished, or where it is useful to efficiently combine several gene variants with small effects.
These markers can also be incorporated in emerging approaches in which breeders are increasingly utilising data from across the whole genome to guide their selections.
A focus on diversity
The project is also looking at phenology in relation to genetic diversity, carrying out a comparison between Australian and global germplasm. One broad aim of these activities is to identify greater variation that might not currently be represented in Australian breeding programs.
A second aim is to follow an alternative approach to unpick the genetic control of phenology through statistical association of trait differences with genes across the genome. This approach is complementary to the more-focused population/progeny approach described above, but has similar benefits with respect to generating markers.
To this end, the project, in collaboration with CSIRO, has assembled a panel of 400 lines from 46 different countries, which broadly captures the global variation in flowering time and genetic diversity and includes 28 accessions released by Australian breeding programs.
This panel is currently being multiplied for subsequent evaluation in the field and under controlled conditions of photoperiod and temperature, which will facilitate the identification of genomic regions regulating flowering in response to each of these components.
Importantly, the panel has also been genotyped using the multi-species pulse DNA chip developed for GRDC by Agriculture Victoria Research. Since this chip is used by pulse breeders and other pre-breeding researchers, it allows researchers to connect pre-breeding research outcomes with the chickpea breeding program, tracking the extent of relevant gene variants within the chickpea germplasm available in Australia.
Testing phenology genes from glasshouse to field
In parallel with tracking down the identities of individual genes, it is also possible to test and validate their effects and interactions, and the project will address this in several ways. One way is to use previously generated populations, and derived progenies that have been specifically selected to represent different combinations of gene variants.
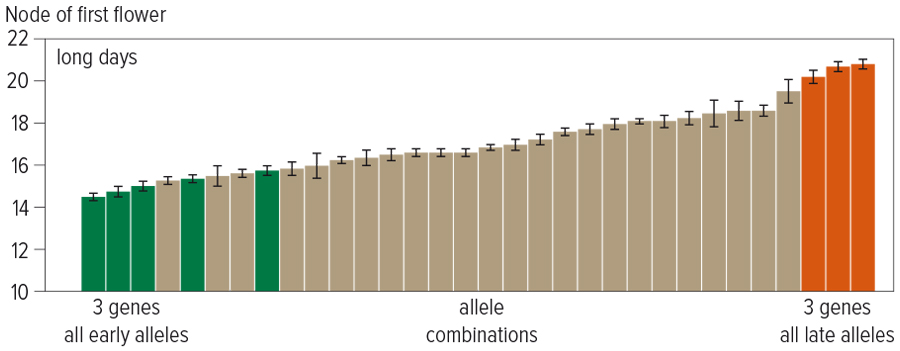
Figure 1: Genes work together to affect flowering time. This graph shows when chickpea plants with different gene combinations start flowering. Green columns have early versions of three genes, yellow columns have later versions and grey columns have a mix of the two sets of genes. Here they have been grown under long daylengths of 16 hours, resulting in a one-week difference, but under short daylengths the difference can be three weeks.
University of Tasmania
For example, the SARDI team has developed a set of closely related lines with different allelic combinations for efl1 and two other flowering genes, and these are being assessed in both field and greenhouse. The results illustrate how variation in phenology can be obtained by combining alleles with distinct effects (Figure 1). Generation of similar sets of lines incorporating other genes is an ongoing activity within the project.
Ultimately this material will be made available to pre-breeders, breeders and agronomists, and will enable testing of the adaptation of different phenology gene combinations across a wider range of environments, including different locations and sowing dates.
More information: Dr Jim Weller, [email protected]

























































