Key points
- Cutting-edge techniques have delivered a more detailed narrow-leafed lupin genome sequence to assist breeders in the development of new varieties
- Development of a pan-genome has harnessed greater diversity of traits for narrow-leafed lupin development
- Mutagenesis has been deployed to create greater trait variation for narrow-leafed lupin.
The Human Genome Project has transformed biology through its unified big science approach to deciphering a reference human genome sequence, along with the complete sequences of key model organisms.
Completed in 2003, it reportedly cost $US3 billion.
Fortunately, the price and quality of sequencing genomes is improving exponentially, now making crop sequencing viable.
Narrow-leafed lupin, a significant legume crop - particularly in Western Australia, where 80 per cent of the world's production is grown - is set to benefit from this type of unified research investment.
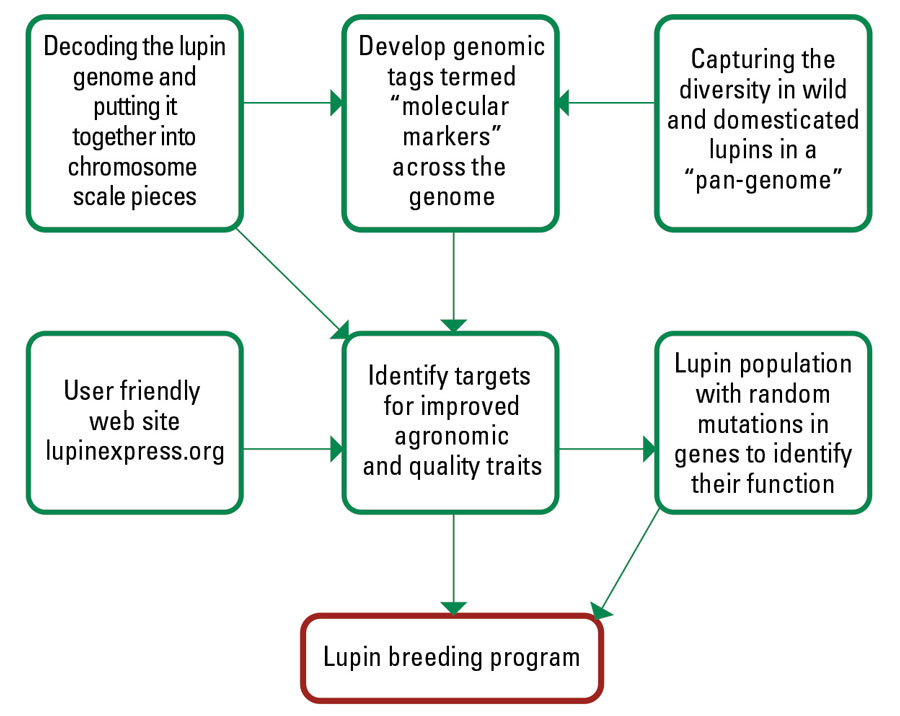
A four-year GRDC investment with CSIRO is providing several tools to help breeders significantly improve traits for lupin growers, producers and consumers (see Figure 1), including:
- decoding the lupin genome sequence;
- generating a pan-genome resource to capture the genetic diversity in narrow-leafed lupins; and
- developing and screening a narrow-leafed lupin mutant population for traits of interest to breeders, growers, and consumers.
Figure 1 Tools developed by the CSIRO team for the lupin research community and lupin breeders.
Source: CSIRO
The first two components of this work are closely linked and provide important input to another GRDC investment led by Dr Sukhjiwan Kaur, at Agriculture Victoria, to generate a pulse breeders' screening tool called a SNP chip to facilitate breeding of key grain legumes in Australia.
SNPs are DNA markers that detect subtle genetic differences in the genome. SNP chips use thousands of these markers to rapidly type genetic diversity within a population. When markers are associated with valuable crop traits, SNP chips allow for the trait's rapid detection and selection within breeding material.
Decoding the lupin genome
An earlier GRDC investment (2012-15, UWA00147) produced and assembled a narrow-leafed lupin genome sequence using what were then state-of-the-art techniques.
However, science has progressed and there is now an opportunity to improve this reference sequence.
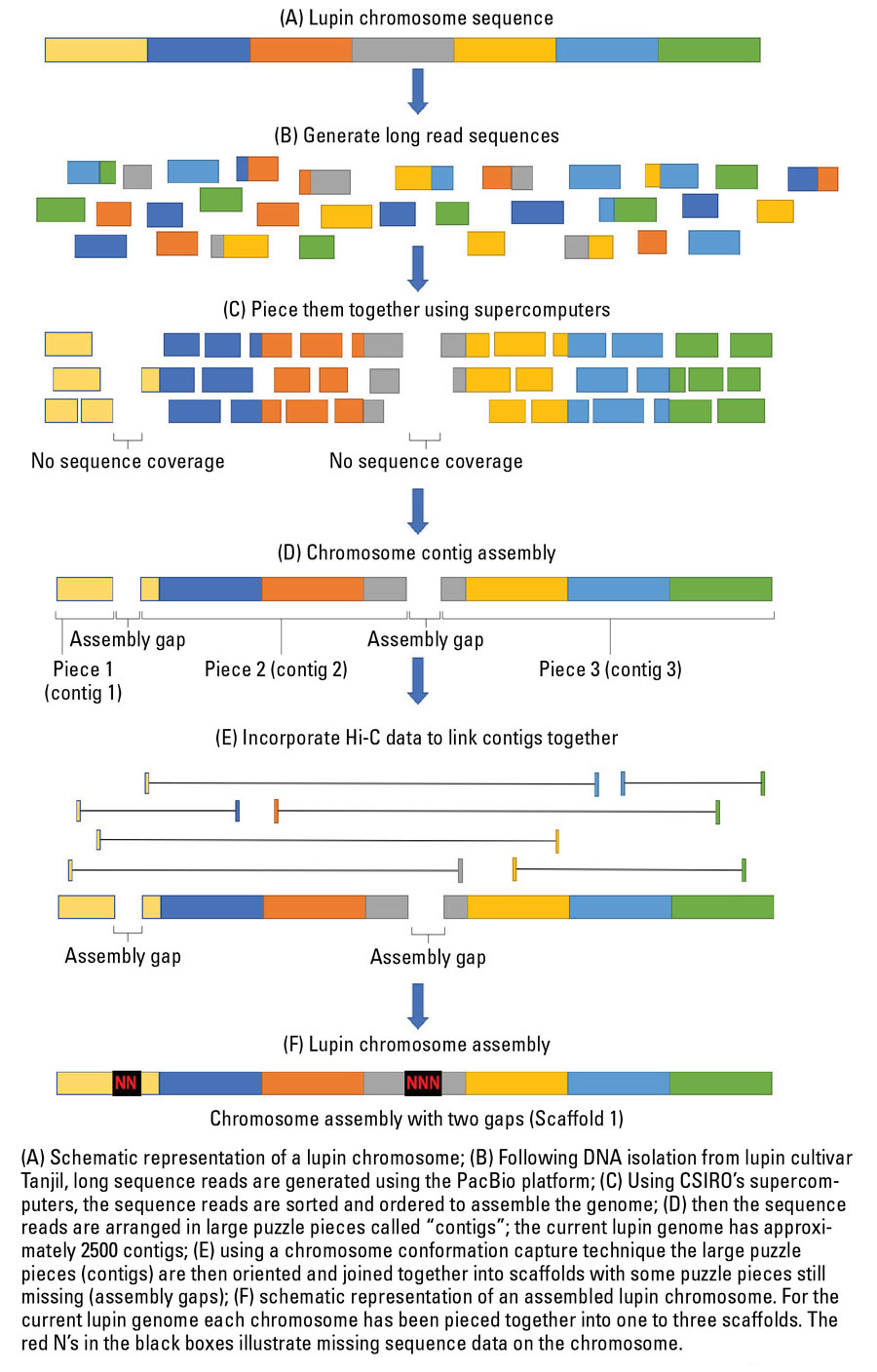
Technical limitations arise because genomes are sequenced by first breaking up the long chromosomal DNA molecule into hundreds of thousands of shorter pieces that can be processed by sequencing technology.
The challenge once the fragments are sequenced is to assemble the sequence into larger pieces (called contigs) before joining up the contigs into scaffolds until the assembled sequence is representative of each chromosome (see Figure 2).
Figure 2 Simplified overview of the lupin genome assembly process on a chromosome scale.
Source: CSIRO
The previous assembly, while capturing essentially all the gene-rich regions, still contained 14,379 scaffolds.
Using improved, long-read sequencing techniques coupled with better assembly methods developed by CSIRO bioinformatician Dr Gagan Garg, the current GRDC investment has produced a less fragmented assembly.
Scaffolds have been reduced to 2136 and, importantly, 90 per cent of the genome is contained in just 24 scaffolds.
These ongoing efforts have resulted in a much better quality narrow-leafed lupin reference genome that is essential to the development of high-throughput genotyping capacity to accelerate narrow-leafed lupin genetic gain.
Lupin pan-genome
The current domesticated narrow-leafed lupin cultivars are derived from a narrow gene pool, resulting in restricted genetic diversity compared with the wealth of diversity found in wild varieties.
To help researchers and breeders tap into this diversity, the CSIRO team is generating a narrow-leafed lupin pan-genome, which will represent the gene set for 20 domesticated varieties and/or breeding lines and 34 wild accessions.
The wild varieties were chosen to be diverse and include material originating from the Iberian Peninsula, north Africa and southern Europe.
They are being compared with domesticated varieties and breeding lines including material of high interest to Australian Grain Technology's lupin breeding program, which works closely with the CSIRO team.
The sequence data from each lupin genome is assembled sequentially against the reference genome to develop a pan-genome assembly.
The price and quality of sequencing genomes is improving exponentially, now allowing vastly improved sequencing of crops to be viable.
While this part of the project is ongoing, its potential to expand the genetic basis of domesticated narrow-leafed lupin is already being explored.
For example, former University of Western Australia PhD student Dr Candy Taylor used pan-genome information alongside mapping information to identify a critical gene involved in the vernalisation response, the trigger for plants to flower once subjected to a cold spell.
Dr Taylor also identified modifications in the gene that allow altered vernalisation traits that could help accelerate uptake of lupin crops in eastern Australia, which has longer growing seasons than the northern wheatbelt of Western Australia.
The pan-genome has also helped identify candidate genes that control traits associated with alkaloid content, pod shattering and disease resistance.
Mutant population traits
TILLING (Targeting Induced Local Lesions IN Genomes) is a 20-year-old method that allows a plant line to be randomly mutated across its genome to create a population with enhanced genetic diversity. The impact of this diversification on genes of interest can then be analysed and exploited.
For example, the beta-conglutins, which are major seed proteins, have been identified as allergens in narrow-leafed lupin by the CSIRO team in collaboration with Sydney University.
Moreover, FSANZ (Food Standards Australia New Zealand) now requires that all lupin processed products must be labelled as containing an allergen. The CSIRO team will look for individual plants from the TILLING population that have mutations in genes encoding specific beta-conglutins, which may result in the allergen(s) being reduced or removed.
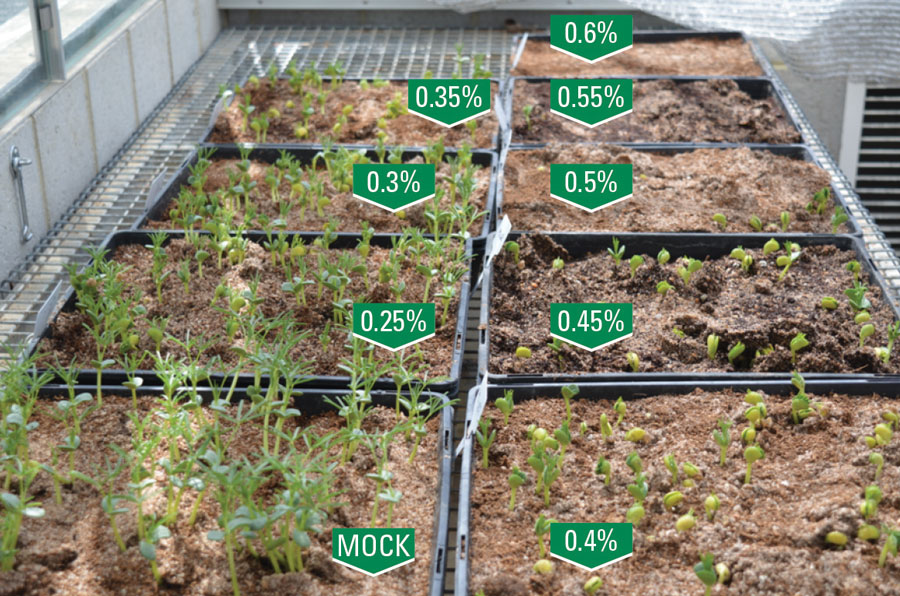
To develop the lupin TILLING population, the CSIRO team tested different concentrations of the chemical that induces mutations. Their goal was a high mutation rate but one that allowed a high percentage of plants to germinate into viable plants (see Figure 3).
Figure 3 Optimising the chemical concentration to produce high mutation rates but maintain plant viability. The tray labelled ‘Mock’ received no chemical treatment while the other trays received the indicated amounts of chemical mutagen.
Source: CSIRO
Genome sequencing is the most thorough way to type the mutated plants. Since the price of sequencing has decreased so much, the team felt it was cost-effective to sequence the whole genome of more than 500 mutant lines. This ensures that all genes are being sequenced, which may identify those that may be of use in the future.
Future varieties
The team has now completed a pilot project in which it sequenced 40 individual mutant narrow-leafed lupin plants and identified 30,487 SNPs (mutations induced by TILLING) in genes that may result in altered traits. Included are 547 SNPs that resulted in a stop-codon that should abolish the gene's function.
This class of mutants is potentially useful for some of the undesirable traits, such as thick seed coat, alkaloid content and allergenicity proteins that are still present in narrow-leafed lupin, given it was only fully domesticated recently (about 60 years ago in Australia).
These tools are central for the future delivery of narrow-leafed lupin varieties.
They allow the identification of new traits that improve cultivation in current growing areas as well as adaptations to changing environments, climates and market demand for the continued prosperity of Australian growers.
More information: Professor Karam Singh, 08 9333 6320, karam.singh@csiro.au

























































