Key points
- Three major genes for blackleg resistance have been identified by the University of Western Australia, doubling the number available to Australian canola breeders
- Researchers at CSIRO have developed new techniques to better identify and characterise the more durable minor gene resistance
The canola industry depends on effective blackleg resistance, but the highly diverse blackleg fungal population is a moving target.
New research has identified three major genes for blackleg resistance and is working to better understand how minor gene resistance is expressed.
Major gene resistance relies on a single gene to confer resistance, while minor gene or ‘quantitative’ resistance depends on a cocktail of minor genes that work together. These two forms of resistance act to control blackleg disease in canola in different ways.
The Australian canola industry has, to date, relied on three major genes, but blackleg populations are highly diverse, with the ability to overcome major genes in as few as three years after commercial release. There is now recognition that a combination of major and minor genes will provide a more durable resistance as it is more difficult for blackleg to overcome multiple genes. However, ‘quantitative’ resistance is difficult to measure in the plant as the effect of each gene on the level of blackleg disease is small and expression is also strongly influenced by the environment, making this a challenging area for development.
Two GRDC investments commenced in 2019 to identify more diverse and effective range of blackleg resistances in canola varieties and to develop reliable methods to phenotype quantitative resistance. The University of Western Australia (UWA) is focusing on major gene resistance, while CSIRO is focusing on minor gene resistance.
New major genes
Since the commencement of this GRDC investment, UWA has doubled the number of major genes identified, which was made possible by combining phenotypic information with state-of-the-art genome-sequencing technologies.
Currently, a range of wild varieties and species are being screened for the identification of novel sources of resistance, while three new genetic sources of resistance are being further characterised.
All current and past Australian varieties were screened to determine and understand which resistance genes they possess. This highlighted that one gene in particular, Rlm4, has been the dominant gene in use by Australian canola breeding companies.
Increasing the number of major genes available to breeders and growers will help avoid the boom-and-bust cycle that results when new resistance genes are quickly overcome. Molecular markers perfectly linked to all six known genes have been developed and are available and can now be used routinely in breeding programs.
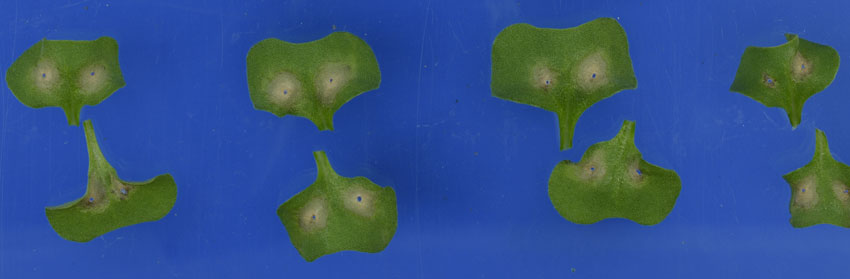
The traditional method of identifying major genes for blackleg resistance involved measuring lesion development on cotyledons after inoculation in the glasshouse. With markers now available for six major genes, this technique can be replaced with a more rapid and reliable screening test of the plant’s DNA. Photo: Dr Susie Sprague
Quantitative resistance
Minor gene or quantitative resistance limits the spread of the fungus once it has infected the plant and is provided by numerous genes each with a small effect.
To date, breeders rely on field assessments of blackleg crown canker severity in mature plants to select for quantitative resistance. However, this strategy depends on the development and expression of disease for visual assessment, which has limitations because:
- the blackleg population is dynamic, changing from year to year;
- the presence of effective major genes masks the presence (or absence) of quantitative resistance genes; and
- there are strong environmental effects that influence disease expression.
Better phenotyping of this type of resistance will make it easier for breeders to identify lines with robust resistance to blackleg.
CSIRO has developed a molecular technique to accurately quantify the amount of blackleg fungus in a plant. This method is being used to identify plant tissues, in which minor gene resistance is acting to limit disease, for targeted phenotyping.
A key finding of the project is that quantitative resistance does not provide the same level of resistance to all blackleg isolates, but instead reacts with individual isolates differently in the plant. This interaction requires further characterisation as it has significant implications for the development of a robust phenotyping method.
More information: Susie Sprague, 0466 643 227, susan.sprague@csiro.au; Jacqueline Batley, 0423 841 669, jacqueline.batley@uwa.edu.au

























































