Like human diseases such as cancer, early detection and monitoring is crucial to keeping crops a step ahead of fungicide resistance.
Achieving this requires continuous monitoring using a combination of sampling, detection and reporting methods that must work efficiently to deliver accurate and timely information to growers to inform practice change (Figure 1).
Figure 1: Workflow underpinning more-agile, in-season fungicide resistance monitoring and practice change.
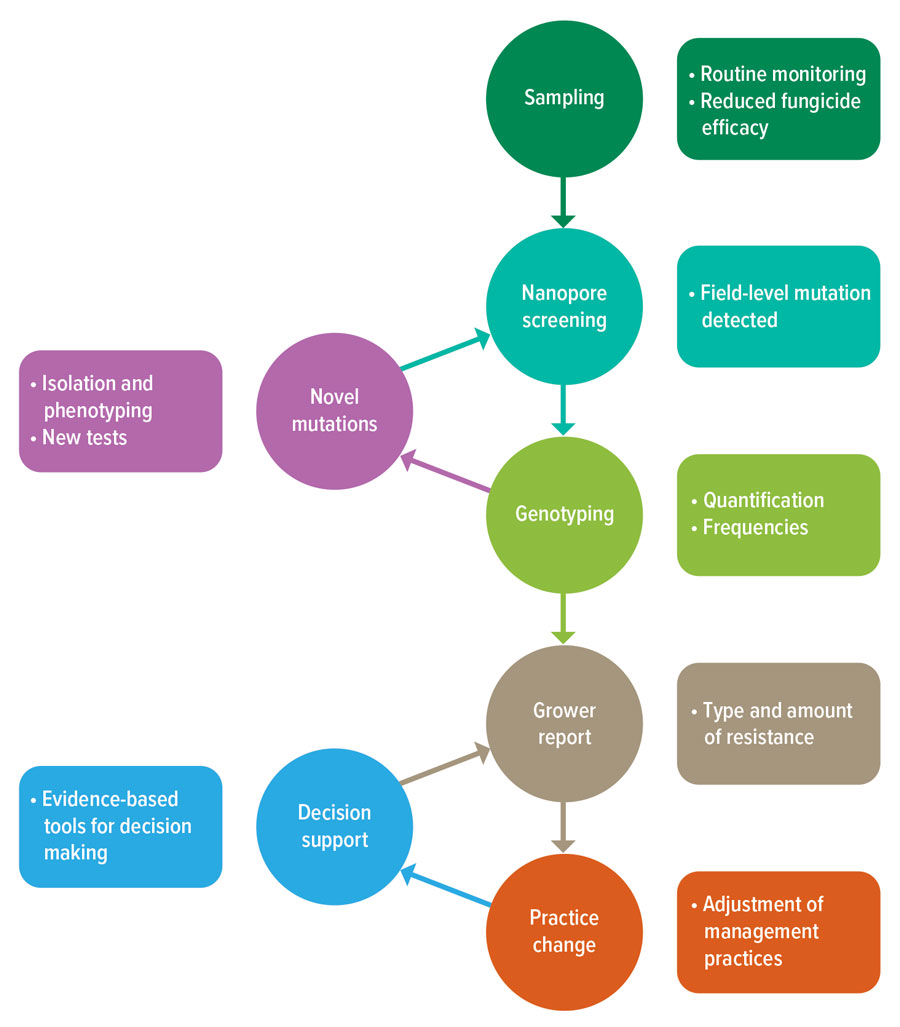
Source: Kat Zulak and Noel Knight
This system must be dynamic and agile. It must also be able to incorporate emerging technologies from fields such as medicine and environmental science to reduce cost, time and resources while maintaining accuracy.
This task means overcoming technical challenges such as sampling bias, detection accuracy, sensitivity and the development of decision-support tools to link laboratory results to implications in the field.
The mission to continuously improve sampling and detection systems is led by the Centre for Crop and Disease Management (CCDM) at Curtin University in Perth. A key objective is to reduce the economic and food security cost of fungicide resistance in Australia.
Sampling
Sampling is the first step in monitoring fungicide resistance in pathogen populations in a crop. Traditionally the detection of fungicide resistance has relied on growers or advisers observing a decrease in fungicide efficacy. Diseased samples, typically from a few positions in afield, are sent to experts to confirm the presence of fungicide resistance.
The quality of the samples can vary depending on the disease, so it is important to consult regional experts. Sampling can also be expanded to examine a larger area of crop along defined transects and include numerous samples. This enables a clearer picture of the frequency of fungicide resistance, and how this might affect disease control.
Proactive, regular field sampling is also important to detect resistance before in-crop failure happens. The detection of fungicide resistance can then be made with phenotyping, genotyping or sequencing.
Phenotyping
Phenotyping is the foundation of resistance detection. It uses living fungi isolated from diseased plant tissues to test the ability of the fungi to survive different fungicide doses. Fungi may be generally classified as sensitive (killed by the fungicide), reduced sensitive (survive low doses of fungicide) or resistant (survive high doses of the fungicide).
A major benefit comes from the ability to link a phenotype to a mutation, or change associated with fungicide resistance, in the target protein and its gene. This leads to a more-precise detection method called genotyping.
Genotyping
Once a mutation is linked to fungicide resistance, genotyping tools can be developed to specifically detect those mutations and quantify them in either fungi grown in the laboratory, or in diseased leaves from the paddock. This is based on the extraction of DNA from the fungi or leaves. Multiple platforms are available for performing DNA detection; however, the development of high-quality tests is needed. Once developed, these analyses take days rather than weeks, increasing the speed of detection compared to the more time-consuming phenotyping.
For each mutation associated with fungicide resistance, a different test must be designed. These tests are sensitive and specific, which means the frequency of resistance in field populations can be accurately quantified.
However, multiple mutations can be present, which can require many tests to be performed. These tests are also limited to already-described mutations. They do not detect new mutations.
Nanopore sequencing
To address the issue of multiple mutations to several fungicides, the CCDM has deployed a sequencing assay using the MinION, a small, portable and cost-effective DNA sequencer, enabling on-demand sequencing and mutation detection. This aims to streamline mutation detection and allow targeted genotyping tests to be performed for fungicide resistance frequency analysis.
However, instead of running multiple genotyping assays on each DNA sample, the target genes are directly sequenced using MinION, revealing known and potentially new mutations simultaneously.
If a new mutation has been detected, the process goes back to the traditional phenotyping method to link the mutation to fungicide resistance.
Recently, researchers have used this method to detect mutations directly from infected leaf samples, enabling large-scale mutation profiling from field samples.
What can growers do?
The principles of managing fungicide resistance rely on integrated management practices to disrupt pathogen survival.
This is based on the selection of the most-resistant crop variety available, integrated disease management such as stubble reduction, crop rotation and controlling green bridges, and finally the need for fungicide applications, which should be strategic and involve the rotation or mixture of different fungicide groups.
Strategic and effective fungicide applications rely on knowledge of the pathogens in the field, and the effect of fungicide resistance on disease control.
As new methods for reporting the presence and frequency of fungicide resistance become available to growers, it will enable more-timely and effective management of fungal diseases.
More information: Kat Zulak, katherine.zulak@curtin.edu.au; Noel Knight, Noel.Knight@unisq.edu.au

























































