Key points
- New developments in DNA testing technology are revolutionising research by identifying disease-causing pathogens in situations where traditional approaches fail and by providing an early warning system through the iMapPESTS surveillance program
- The technology is also being adapted to provide a DNA test to confirm the presence of group E and F rhizobia prior to sowing pulses
Accurate identification of fungal disease pests and plant rhizobium will help both researchers and growers make better decisions.
Every living organism has its own unique genetic code. Now, DNA testing methods are overtaking traditional processes to more accurately identify organisms that impact on agriculture.
Traditional methods of identifying disease-causing pathogens involve detailed visual assessments or time-consuming processes of growing the pathogen to a more identifiable stage in the life cycle.
Researchers at the Department of Primary Industries and Regions research division South Australian Research and Development Institute’s (SARDI) Molecular Diagnostic Centre (MDC) are breaking down these barriers with new DNA tests.
This capability is supporting a diverse range of projects from disease identification and monitoring through to the development of a new range of tests to quantify rhizobial populations in soils.
Soil-borne disease identification
Some important pathogens are difficult to isolate, which is frustrating for both growers and researchers.
A classic example was the identification of the pathogens to explain the severe symptoms associated with several chickpea crop failures in South Australia in 2017. This triggered a series of GRDC and South Australian Grain Industry Trust (SAGIT) projects to investigate soil-borne diseases of pulses and oilseeds nationally.

Next-generation sequencing methods developed by SARDI’s Molecular Diagnosis Centre played a key role in identifying pathogens associated with severe disease in several chickpea crops in SA in 2017. Photo: Adam Hancock, Elders
To support this research, the MDC used next-generation sequencing methods to identify potentially important fungal pathogens including oomycetes (such as Phytophthora), and Fusarium species that could be easily missed using pathogen isolation techniques.
The MDC analysed DNA extracted from diseased chickpea and faba bean crops near where the original crop failures occurred and identified multiple root rot pathogens (Phytophthora, Aphanomyces, black root rot and a number of Fusarium sp.).
Of surprise was a number of major root pathogens reported elsewhere in the world that were already present in Australian pulse crops.
Phytophthora root rot – a serious problem for chickpeas in the northern region – was known to be caused by Phytophthora medicaginis. However, this research identified several other disease-causing Phytophthora species in chickpea, faba bean, lentil and lupin crops across the country.
While the Phytophthora story is interesting, Fusarium and possibly Phoma species might turn out to be more important in Australia as new information is discovered.
Fusarium species that are pathogenic on faba beans, peas, lentils and chickpeas – Sclerotinia trifoliorum on legume crops and Phytophthora drechsleri on lupins – have also been found associated with disease. This information brings us closer to finding the causative agent(s) of these root disease observations including the 2017 event and will help inform future research.
Early warning system
SARDI’s role in a Rural R&D for Profit project, iMapPESTS, includes developing spore-trapping technology to inform growers when crops are likely to be exposed to infection by specific airborne pathogens. The project’s ‘sentinels’ provide platforms to monitor and report the presence of airborne pests and diseases affecting major agricultural sectors nationwide, including grains, cotton, sugar, horticulture, viticulture and forestry.

SARDI’s Dr Rohan Kimber says DNA testing from the iMapPESTS spore traps is revealing new information about pathogen behaviour. Photo: Andrew Beveridge
The project uses DNA testing to objectively and accurately track changes in spore populations over time, often triggered by seasonal conditions and farming systems.
Once established, timely information on pathogen abundance and spread will improve pest management decision-making, biosecurity responses to exotic pests and diseases, and support surveys and area freedom claims.
DNA testing from spore traps is revealing new information about pathogen behaviour. Deeper analysis of pathogen dispersal events with weather data aims to identify key triggers in the growing season to inform pathogen risk profiles.
Growers can use the live results from this new-generation surveillance technology to guide scouting efforts and pest control action.
Identifying rhizobia
The value of the DNA tools is not just restricted to disease-causing organisms. Growers have long desired a test to determine whether rhizobial inoculants are required when sowing pulses in paddocks with a previous history of the crop. But existing methods used to study rhizobial populations in the field have proved too laborious to use as the basis of a grower service.
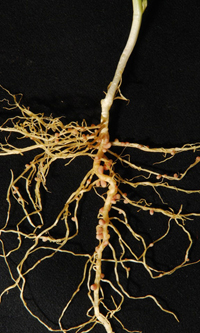
While traditionally focused on disease testing, the Molecular Diagnostic Centre has now developed a DNA test for group E and F rhizobia, which nodulate field peas, lentils, faba beans and vetch. Photo: Dr Liz Farquharson
The test was released to southern region growers in February 2021 as ‘PREDICTA® rNod’ and will be delivered nationally from 2022. As much as it will be welcomed by growers, this new technology will have an even greater impact on field research.
The MDC has been working to develop a DNA test since 2013 (DAS00115 and DAS00137), but it took until 2019 to make the breakthrough that led to the successful development of a test for group E and F rhizobia, Rhizobium leguminosarum bv. viciae. The test is extremely sensitive and, importantly, does not detect the closely related clover rhizobia (R. leguminosarum bv. trifolii).
Further research is underway to develop tests for the rhizobia associated with chickpeas, lupins and mungbeans.
More information: Dr Alan McKay, 08 8429 2216, alan.mckay@sardi.gov.au; iMapPESTS

























































